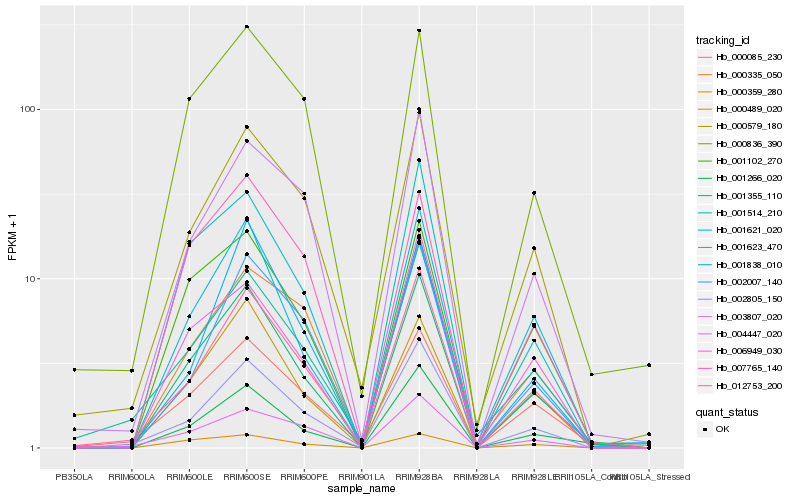
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001355_110 |
0.0 |
- |
- |
PREDICTED: mitochondrial chaperone BCS1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_001266_020 |
0.0836350571 |
- |
- |
PREDICTED: uncharacterized protein LOC105650395 [Jatropha curcas] |
| 3 |
Hb_001623_470 |
0.0905009646 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 4 |
Hb_001621_020 |
0.090573381 |
- |
- |
- |
| 5 |
Hb_001102_270 |
0.09471336 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 6 |
Hb_012753_200 |
0.097743406 |
- |
- |
hypothetical protein JCGZ_20651 [Jatropha curcas] |
| 7 |
Hb_000489_020 |
0.1011267398 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 8 |
Hb_002805_150 |
0.1070008063 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 9 |
Hb_000579_180 |
0.1233585706 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 10 |
Hb_007765_140 |
0.1236168205 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 8.1-like [Jatropha curcas] |
| 11 |
Hb_003807_020 |
0.1237386462 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 12 |
Hb_006949_030 |
0.1239125702 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000085_230 |
0.1244300582 |
- |
- |
PREDICTED: probable long-chain-alcohol O-fatty-acyltransferase 5 [Jatropha curcas] |
| 14 |
Hb_000836_390 |
0.1285094657 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 15 |
Hb_000335_050 |
0.1295078914 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000359_280 |
0.1304245795 |
- |
- |
hypothetical protein OsI_15300 [Oryza sativa Indica Group] |
| 17 |
Hb_001514_210 |
0.1319920187 |
- |
- |
hypothetical protein VITISV_012133 [Vitis vinifera] |
| 18 |
Hb_002007_140 |
0.1331193336 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like [Populus euphratica] |
| 19 |
Hb_004447_020 |
0.1333490507 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 20 |
Hb_001838_010 |
0.1351839525 |
- |
- |
hypothetical protein POPTR_0005s18380g [Populus trichocarpa] |