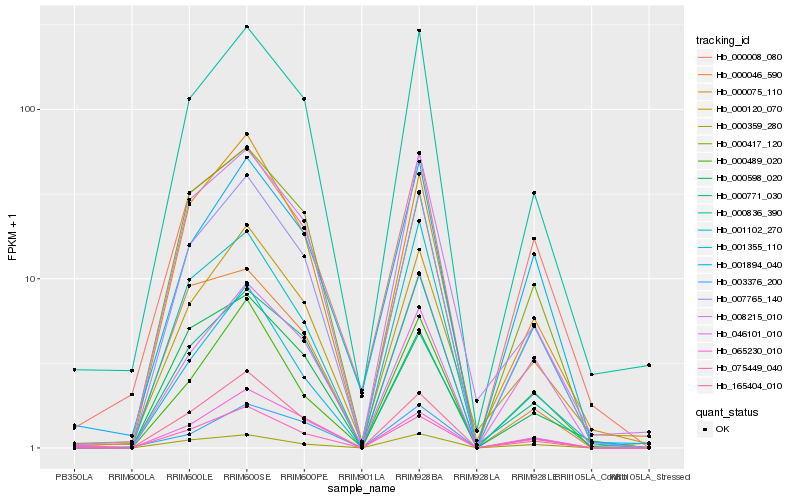
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007765_140 |
0.0 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 8.1-like [Jatropha curcas] |
| 2 |
Hb_000836_390 |
0.0696103661 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 3 |
Hb_165404_010 |
0.0724784696 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 4 |
Hb_000075_110 |
0.0742650212 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 5 |
Hb_075449_040 |
0.0795748917 |
- |
- |
PREDICTED: meiotic recombination protein SPO11-1 [Jatropha curcas] |
| 6 |
Hb_008215_010 |
0.0841215825 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000120_070 |
0.0852569291 |
- |
- |
PREDICTED: uncharacterized protein LOC105630351 [Jatropha curcas] |
| 8 |
Hb_065230_010 |
0.1046757163 |
- |
- |
PREDICTED: uncharacterized protein LOC100814166 isoform X1 [Glycine max] |
| 9 |
Hb_001102_270 |
0.1075448564 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 10 |
Hb_000359_280 |
0.107960507 |
- |
- |
hypothetical protein OsI_15300 [Oryza sativa Indica Group] |
| 11 |
Hb_000489_020 |
0.1169456743 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 12 |
Hb_000417_120 |
0.1173931459 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 13 |
Hb_001894_040 |
0.1186789471 |
- |
- |
PREDICTED: carbonic anhydrase 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000598_020 |
0.1201267881 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 15 |
Hb_003376_200 |
0.1219836816 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_046101_010 |
0.1228926498 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 17 |
Hb_001355_110 |
0.1236168205 |
- |
- |
PREDICTED: mitochondrial chaperone BCS1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000771_030 |
0.1236812893 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000008_080 |
0.1273953783 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000046_590 |
0.1280411319 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |