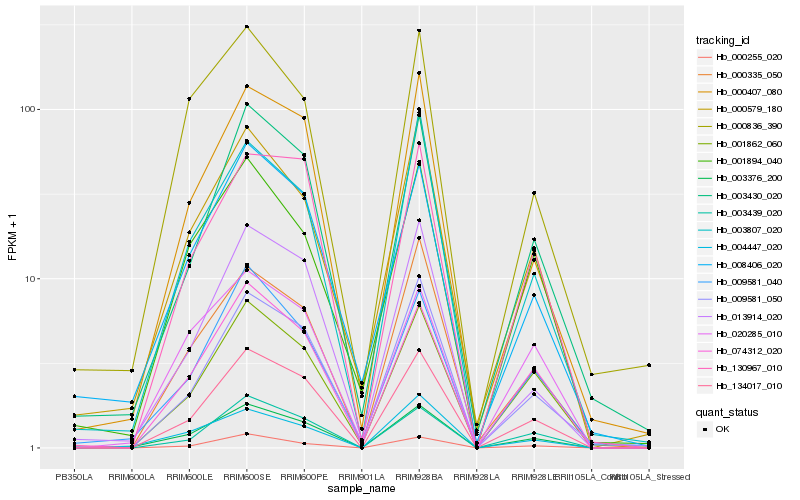
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003376_200 |
0.0 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_134017_010 |
0.0757846848 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_074312_020 |
0.095110314 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_004447_020 |
0.0976388463 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 5 |
Hb_001862_060 |
0.1021842027 |
- |
- |
PREDICTED: uncharacterized protein LOC105639802 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000407_080 |
0.1031289746 |
- |
- |
1-aminocyclopropane-1-carboxylate oxidase [Hevea brasiliensis] |
| 7 |
Hb_008406_020 |
0.1076426674 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 8 |
Hb_009581_040 |
0.1079022926 |
- |
- |
hypothetical protein B456_011G052100 [Gossypium raimondii] |
| 9 |
Hb_000579_180 |
0.1085553213 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 10 |
Hb_020285_010 |
0.1094092752 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3, partial [Pyrus x bretschneideri] |
| 11 |
Hb_000836_390 |
0.1122449053 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 12 |
Hb_003430_020 |
0.1122712169 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_000255_020 |
0.1123611148 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 14 |
Hb_003439_020 |
0.1134944601 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1-like [Solanum lycopersicum] |
| 15 |
Hb_000335_050 |
0.114477915 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001894_040 |
0.1167680901 |
- |
- |
PREDICTED: carbonic anhydrase 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003807_020 |
0.1169294673 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 18 |
Hb_130967_010 |
0.1183841615 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_009581_050 |
0.120840851 |
- |
- |
hypothetical protein CISIN_1g0025812mg, partial [Citrus sinensis] |
| 20 |
Hb_013914_020 |
0.1216815478 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |