| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
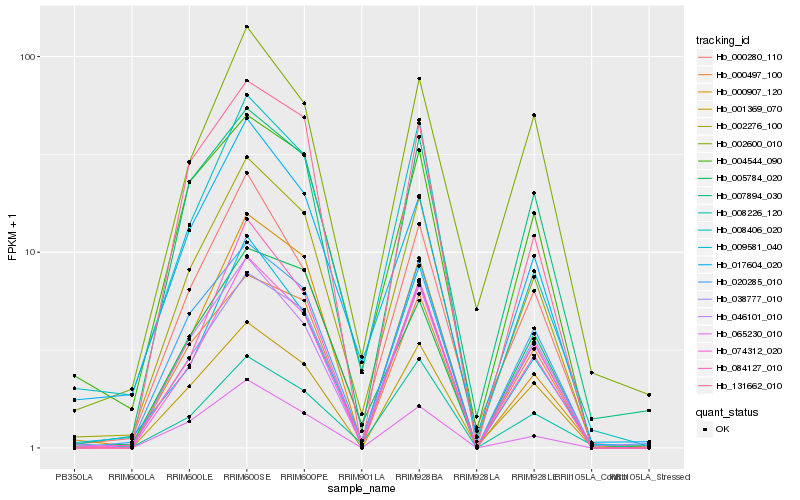
Hb_002276_100 |
0.0 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89A2-like [Jatropha curcas] |
| 2 |
Hb_074312_020 |
0.082444942 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_020285_010 |
0.0828376143 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3, partial [Pyrus x bretschneideri] |
| 4 |
Hb_000280_110 |
0.0864207924 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 5 |
Hb_046101_010 |
0.0886156592 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 6 |
Hb_038777_010 |
0.0924852057 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN2 isoform X1 [Populus euphratica] |
| 7 |
Hb_001369_070 |
0.0949504584 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_000497_100 |
0.0959838548 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 9 |
Hb_004544_090 |
0.0960311931 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_008406_020 |
0.0964139581 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 11 |
Hb_084127_010 |
0.0981962946 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_009581_040 |
0.1001411621 |
- |
- |
hypothetical protein B456_011G052100 [Gossypium raimondii] |
| 13 |
Hb_017604_020 |
0.1016630857 |
- |
- |
hypothetical protein JCGZ_22673 [Jatropha curcas] |
| 14 |
Hb_005784_020 |
0.1030777253 |
- |
- |
Protein MKS1, putative [Ricinus communis] |
| 15 |
Hb_007894_030 |
0.1032114751 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 16 |
Hb_000907_120 |
0.1077574064 |
transcription factor |
TF Family: G2-like |
PREDICTED: transcription repressor KAN1-like [Populus euphratica] |
| 17 |
Hb_065230_010 |
0.1099695824 |
- |
- |
PREDICTED: uncharacterized protein LOC100814166 isoform X1 [Glycine max] |
| 18 |
Hb_002600_010 |
0.1101343073 |
- |
- |
Allene oxide cyclase 4, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_008226_120 |
0.1111310698 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_131662_010 |
0.1146637608 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |