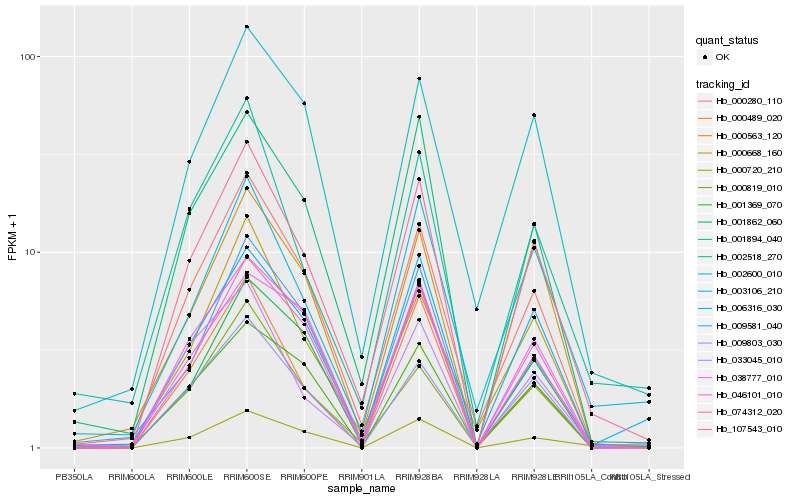
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_107543_010 |
0.0 |
- |
- |
Cysteine-rich RLK 10, putative [Theobroma cacao] |
| 2 |
Hb_000280_110 |
0.0927584655 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 3 |
Hb_046101_010 |
0.1034571591 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 4 |
Hb_001894_040 |
0.1039492128 |
- |
- |
PREDICTED: carbonic anhydrase 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_009803_030 |
0.118337866 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 6 |
Hb_000489_020 |
0.1216598322 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 7 |
Hb_074312_020 |
0.1222109814 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_006316_030 |
0.1229583443 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 9 |
Hb_000668_160 |
0.1234460791 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 10 |
Hb_033045_010 |
0.1246898924 |
- |
- |
copine, putative [Ricinus communis] |
| 11 |
Hb_009581_040 |
0.125182838 |
- |
- |
hypothetical protein B456_011G052100 [Gossypium raimondii] |
| 12 |
Hb_000819_010 |
0.1258029828 |
- |
- |
Receptor-like protein kinase 1, putative [Theobroma cacao] |
| 13 |
Hb_002600_010 |
0.1263945348 |
- |
- |
Allene oxide cyclase 4, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_001369_070 |
0.1270608233 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000720_210 |
0.1276299871 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 16 |
Hb_000563_120 |
0.1296388643 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 17 |
Hb_001862_060 |
0.1323981136 |
- |
- |
PREDICTED: uncharacterized protein LOC105639802 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002518_270 |
0.1341279979 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 19 |
Hb_038777_010 |
0.1343481242 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN2 isoform X1 [Populus euphratica] |
| 20 |
Hb_003106_210 |
0.1362869971 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74B1-like [Jatropha curcas] |