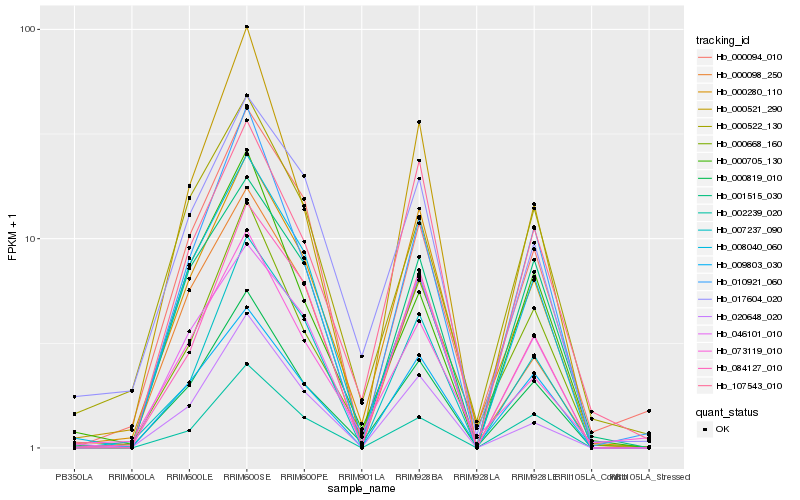
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000819_010 |
0.0 |
- |
- |
Receptor-like protein kinase 1, putative [Theobroma cacao] |
| 2 |
Hb_000668_160 |
0.0826219274 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 3 |
Hb_009803_030 |
0.0858279259 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 4 |
Hb_000280_110 |
0.0873530779 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 5 |
Hb_010921_060 |
0.0922857343 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_008040_060 |
0.1027511541 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 7 |
Hb_000522_130 |
0.1080704941 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 8 |
Hb_073119_010 |
0.1088331525 |
- |
- |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 9 |
Hb_000521_290 |
0.113140013 |
- |
- |
hypothetical protein POPTR_0001s23530g [Populus trichocarpa] |
| 10 |
Hb_017604_020 |
0.1150117134 |
- |
- |
hypothetical protein JCGZ_22673 [Jatropha curcas] |
| 11 |
Hb_000705_130 |
0.1151784753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002239_020 |
0.1165865076 |
- |
- |
MLO, putative [Theobroma cacao] |
| 13 |
Hb_000094_010 |
0.1226729113 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 14 |
Hb_001515_030 |
0.1233033459 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_020648_020 |
0.1244970781 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor pho1, putative [Ricinus communis] |
| 16 |
Hb_107543_010 |
0.1258029828 |
- |
- |
Cysteine-rich RLK 10, putative [Theobroma cacao] |
| 17 |
Hb_084127_010 |
0.1288534549 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_000098_250 |
0.1289916496 |
- |
- |
UDP-glucose flavonoid 3-O-glucosyltransferase [Medicago truncatula] |
| 19 |
Hb_007237_090 |
0.1292657561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_046101_010 |
0.1307470759 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |