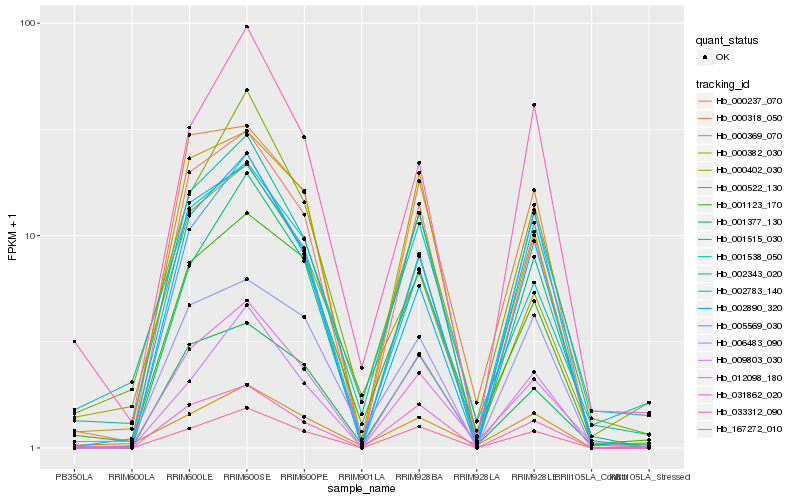
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001538_050 |
0.0 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 2 |
Hb_012098_180 |
0.0906766585 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 3 |
Hb_000522_130 |
0.10136933 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 4 |
Hb_000237_070 |
0.1021898822 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 5 |
Hb_001123_170 |
0.1114131296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001377_130 |
0.1122008111 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 7 |
Hb_000382_030 |
0.1139652664 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 8 |
Hb_167272_010 |
0.1145272362 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 9 |
Hb_001515_030 |
0.1160162689 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_033312_090 |
0.1170452021 |
- |
- |
PREDICTED: HMG-Y-related protein A-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_031862_020 |
0.1209767735 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 12 |
Hb_002343_020 |
0.1213339851 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 13 |
Hb_002783_140 |
0.1215634295 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 14 |
Hb_000369_070 |
0.122814735 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 15 |
Hb_006483_090 |
0.1233497924 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 16 |
Hb_000402_030 |
0.1256554321 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 17 |
Hb_000318_050 |
0.1259170008 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 18 |
Hb_009803_030 |
0.1261123228 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 19 |
Hb_005569_030 |
0.1276067272 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002890_320 |
0.1300428807 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |