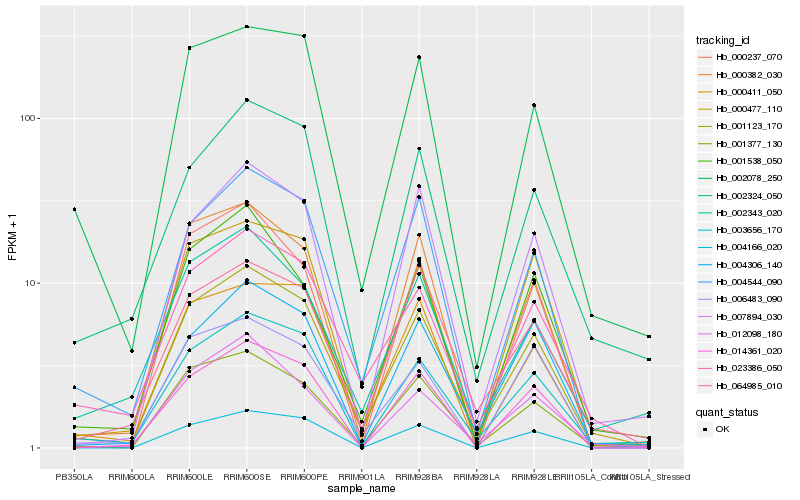
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001123_170 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003656_170 |
0.0745303057 |
- |
- |
PREDICTED: phenylalanine ammonia-lyase-like [Jatropha curcas] |
| 3 |
Hb_001377_130 |
0.0817069653 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 4 |
Hb_004544_090 |
0.0861010196 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_000382_030 |
0.0939290427 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 6 |
Hb_000477_110 |
0.103213504 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 7 |
Hb_002078_250 |
0.1036093624 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004306_140 |
0.1040262173 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 9 |
Hb_023386_050 |
0.104128391 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007894_030 |
0.1046082833 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 11 |
Hb_000411_050 |
0.1101989488 |
- |
- |
hypothetical protein AMTR_s00066p00166230 [Amborella trichopoda] |
| 12 |
Hb_001538_050 |
0.1114131296 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 13 |
Hb_014361_020 |
0.1122245745 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 14 |
Hb_002324_050 |
0.1133008659 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 15 |
Hb_012098_180 |
0.1139341343 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 16 |
Hb_002343_020 |
0.1142018649 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 17 |
Hb_004166_020 |
0.1177456057 |
- |
- |
- |
| 18 |
Hb_064985_010 |
0.1189215933 |
- |
- |
Leucine-rich repeat transmembrane protein kinase, putative [Theobroma cacao] |
| 19 |
Hb_006483_090 |
0.1202393082 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 20 |
Hb_000237_070 |
0.1219029514 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |