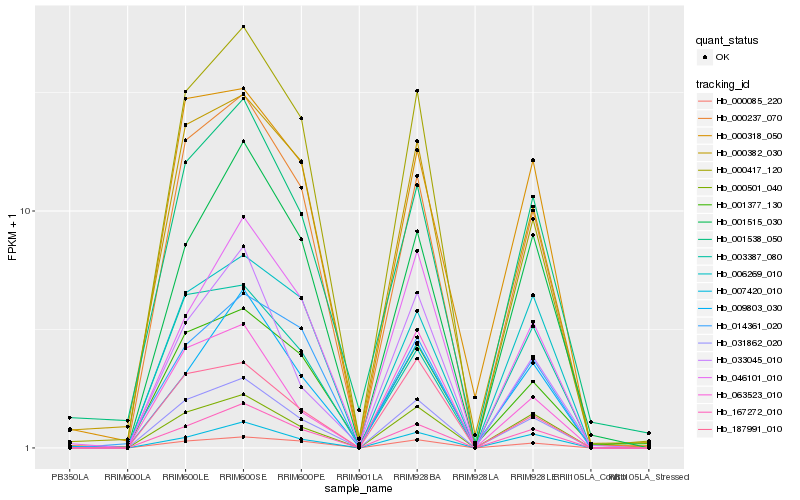
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031862_020 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 2 |
Hb_167272_010 |
0.0701833893 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 3 |
Hb_000237_070 |
0.0724513177 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 4 |
Hb_000501_040 |
0.0741413121 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_000085_220 |
0.0882735774 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000382_030 |
0.0928933726 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 7 |
Hb_007420_010 |
0.1021393132 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X2 [Jatropha curcas] |
| 8 |
Hb_187991_010 |
0.107347143 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 9 |
Hb_000318_050 |
0.1125612555 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001377_130 |
0.1163359472 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 11 |
Hb_000417_120 |
0.1179952438 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 12 |
Hb_063523_010 |
0.1186042333 |
- |
- |
- |
| 13 |
Hb_001538_050 |
0.1209767735 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 14 |
Hb_001515_030 |
0.1226227913 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_006269_010 |
0.1233974426 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 16 |
Hb_014361_020 |
0.1234127407 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 17 |
Hb_009803_030 |
0.1241591192 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 18 |
Hb_046101_010 |
0.1243830381 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 19 |
Hb_033045_010 |
0.1246187758 |
- |
- |
copine, putative [Ricinus communis] |
| 20 |
Hb_003387_080 |
0.1266625156 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |