| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
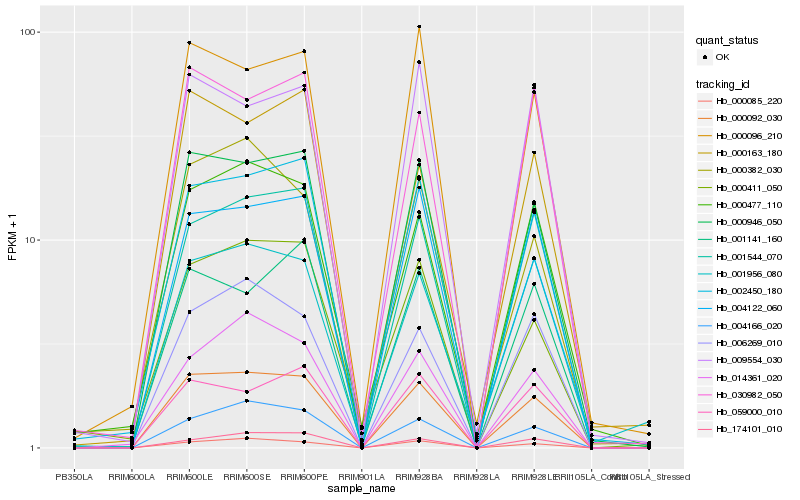
Hb_000092_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105111213 [Populus euphratica] |
| 2 |
Hb_000946_050 |
0.0451981033 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_000085_220 |
0.0795099655 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004166_020 |
0.0860013081 |
- |
- |
- |
| 5 |
Hb_001544_070 |
0.0887638152 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 6 |
Hb_004122_060 |
0.0919966953 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280-like [Jatropha curcas] |
| 7 |
Hb_006269_010 |
0.0972933271 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 8 |
Hb_000477_110 |
0.1054602855 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 9 |
Hb_030982_050 |
0.1056509163 |
- |
- |
PREDICTED: gamma-glutamyl hydrolase 2-like [Jatropha curcas] |
| 10 |
Hb_000096_210 |
0.1057289745 |
- |
- |
PREDICTED: probable carboxylesterase 7 [Jatropha curcas] |
| 11 |
Hb_059000_010 |
0.107531797 |
- |
- |
hypothetical protein JCGZ_17736 [Jatropha curcas] |
| 12 |
Hb_014361_020 |
0.1077259282 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 13 |
Hb_002450_180 |
0.1108124686 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_174101_010 |
0.1117467112 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 15 |
Hb_000382_030 |
0.1159640016 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 16 |
Hb_001141_160 |
0.1161844508 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 17 |
Hb_001956_080 |
0.1175019742 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 18 |
Hb_000411_050 |
0.1210978725 |
- |
- |
hypothetical protein AMTR_s00066p00166230 [Amborella trichopoda] |
| 19 |
Hb_009554_030 |
0.1212265519 |
- |
- |
PREDICTED: chlorophyll synthase, chloroplastic [Prunus mume] |
| 20 |
Hb_000163_180 |
0.1230038636 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |