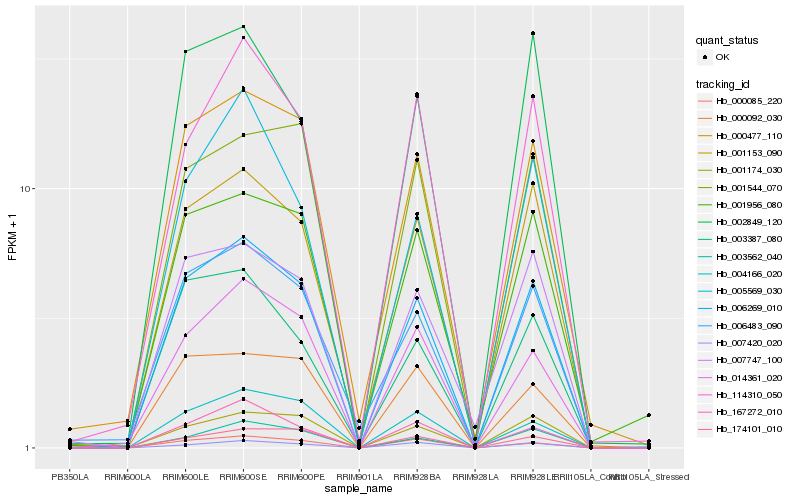
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006269_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 2 |
Hb_001153_090 |
0.0735080648 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 3 |
Hb_001174_030 |
0.0845355901 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_000477_110 |
0.0881195711 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 5 |
Hb_000085_220 |
0.0910805793 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_014361_020 |
0.092469097 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 7 |
Hb_004166_020 |
0.0932679602 |
- |
- |
- |
| 8 |
Hb_167272_010 |
0.0963361458 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 9 |
Hb_000092_030 |
0.0972933271 |
- |
- |
PREDICTED: uncharacterized protein LOC105111213 [Populus euphratica] |
| 10 |
Hb_001544_070 |
0.1000602708 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 11 |
Hb_003387_080 |
0.1008009142 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_007747_100 |
0.1011089553 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 13 |
Hb_006483_090 |
0.1016549195 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 14 |
Hb_001956_080 |
0.1030775121 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 15 |
Hb_114310_050 |
0.1031556855 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 16 |
Hb_003562_040 |
0.1043306286 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1-like [Jatropha curcas] |
| 17 |
Hb_174101_010 |
0.1047750629 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 18 |
Hb_005569_030 |
0.1055618359 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002849_120 |
0.106019606 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 20 |
Hb_007420_020 |
0.1078174131 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |