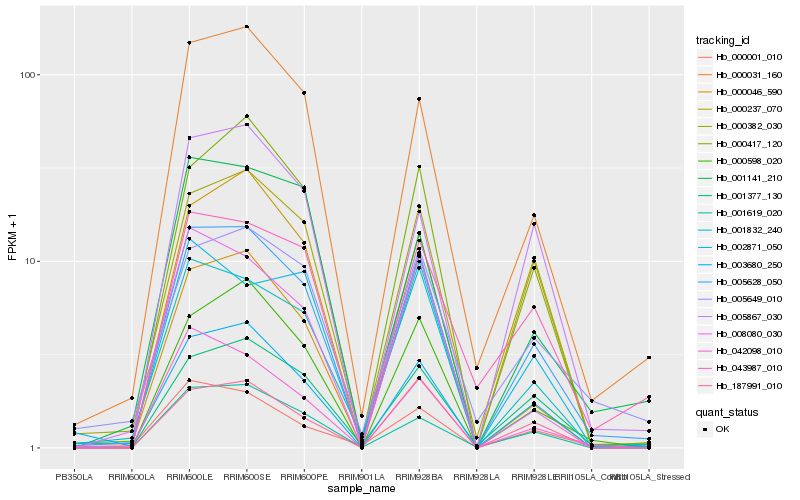
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005628_050 |
0.0 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 2 |
Hb_000031_160 |
0.1036569335 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 3 |
Hb_001832_240 |
0.1049719822 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 4 |
Hb_000382_030 |
0.105325699 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 5 |
Hb_001377_130 |
0.1116714346 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 6 |
Hb_000598_020 |
0.112761834 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 7 |
Hb_000417_120 |
0.1142310755 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 8 |
Hb_003680_250 |
0.1174716056 |
- |
- |
PREDICTED: probable 3-deoxy-D-manno-octulosonic acid transferase, mitochondrial isoform X2 [Jatropha curcas] |
| 9 |
Hb_001619_020 |
0.1180520796 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 10 |
Hb_000001_010 |
0.121874699 |
- |
- |
PREDICTED: uncharacterized protein LOC105648961 [Jatropha curcas] |
| 11 |
Hb_042098_010 |
0.1290761746 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 12 |
Hb_000046_590 |
0.1290944596 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 13 |
Hb_000237_070 |
0.1326170977 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 14 |
Hb_005649_010 |
0.1330706391 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 15 |
Hb_005867_030 |
0.1335139602 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 16 |
Hb_187991_010 |
0.1337699842 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 17 |
Hb_043987_010 |
0.138497539 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 18 |
Hb_008080_030 |
0.1396378265 |
- |
- |
hypothetical protein POPTR_0275s002101g, partial [Populus trichocarpa] |
| 19 |
Hb_001141_210 |
0.1443689435 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 20 |
Hb_002871_050 |
0.1446001117 |
transcription factor |
TF Family: ARF |
auxin response factor 1 family protein [Populus trichocarpa] |