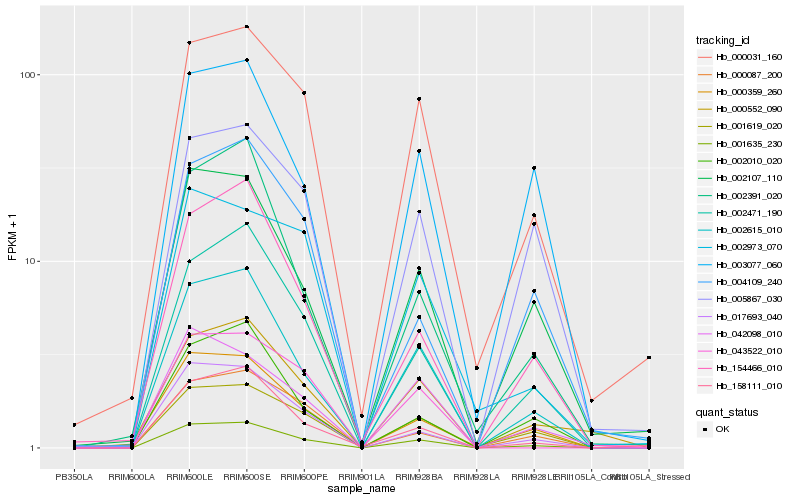
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001635_230 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_002615_010 |
0.0658006361 |
- |
- |
Uncharacterized protein TCM_040942 [Theobroma cacao] |
| 3 |
Hb_002471_190 |
0.0918353815 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 4 |
Hb_017693_040 |
0.1009492987 |
- |
- |
tyrosine kinase family protein [Medicago truncatula] |
| 5 |
Hb_000359_260 |
0.1037740314 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 6 |
Hb_001619_020 |
0.1072723462 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 7 |
Hb_158111_010 |
0.1137337555 |
- |
- |
S-locus-specific glycoprotein precursor, putative [Ricinus communis] |
| 8 |
Hb_002107_110 |
0.11415404 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 6 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000031_160 |
0.1151385596 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 10 |
Hb_154466_010 |
0.1199961063 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 11 |
Hb_000087_200 |
0.1207287345 |
- |
- |
PREDICTED: uncharacterized protein LOC105641954 [Jatropha curcas] |
| 12 |
Hb_002010_020 |
0.1217117511 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 13 |
Hb_000552_090 |
0.1280050213 |
- |
- |
hypothetical protein JCGZ_18511 [Jatropha curcas] |
| 14 |
Hb_042098_010 |
0.1326016693 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 15 |
Hb_002391_020 |
0.1356130079 |
transcription factor |
TF Family: NAC |
PREDICTED: putative NAC domain-containing protein 94 [Jatropha curcas] |
| 16 |
Hb_003077_060 |
0.1384249863 |
- |
- |
PREDICTED: 3-oxo-Delta(4,5)-steroid 5-beta-reductase-like [Jatropha curcas] |
| 17 |
Hb_005867_030 |
0.1431598737 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 18 |
Hb_043522_010 |
0.1438775284 |
- |
- |
- |
| 19 |
Hb_004109_240 |
0.1479612417 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 20 |
Hb_002973_070 |
0.1520731719 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |