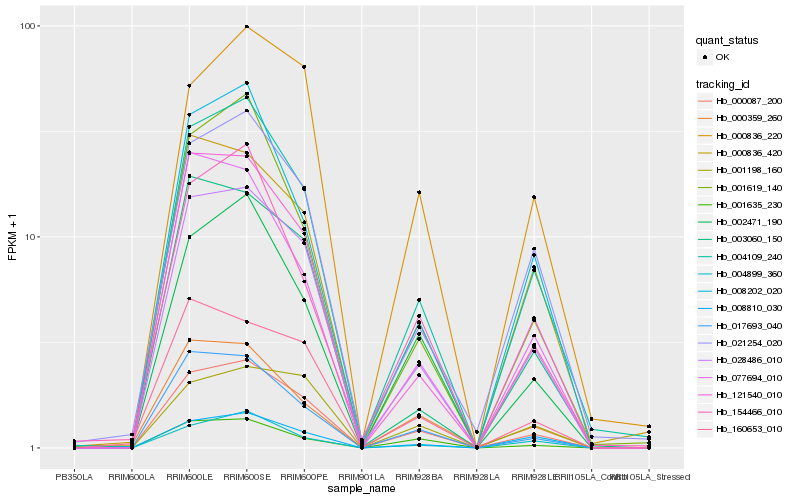
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000087_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641954 [Jatropha curcas] |
| 2 |
Hb_028486_010 |
0.0485482896 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 3 |
Hb_004109_240 |
0.0724317867 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 4 |
Hb_002471_190 |
0.0934796273 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 5 |
Hb_017693_040 |
0.0976528059 |
- |
- |
tyrosine kinase family protein [Medicago truncatula] |
| 6 |
Hb_121540_010 |
0.0978775906 |
- |
- |
PREDICTED: UDP-glycosyltransferase 88A1-like [Jatropha curcas] |
| 7 |
Hb_021254_020 |
0.1125452357 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 8 |
Hb_160653_010 |
0.1184966691 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 9 |
Hb_001635_230 |
0.1207287345 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_000836_420 |
0.1220393788 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 11 |
Hb_008810_030 |
0.1251151013 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_001619_140 |
0.1257546345 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 13 |
Hb_154466_010 |
0.1272248411 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 14 |
Hb_001198_160 |
0.1274232617 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 15 |
Hb_004899_360 |
0.1278959464 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 16 |
Hb_000836_220 |
0.1282108791 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_003060_150 |
0.1284483272 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_008202_020 |
0.1311215368 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 19 |
Hb_000359_260 |
0.1328042671 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 20 |
Hb_077694_010 |
0.1362150413 |
- |
- |
uncharacterized protein LOC100305624 [Glycine max] |