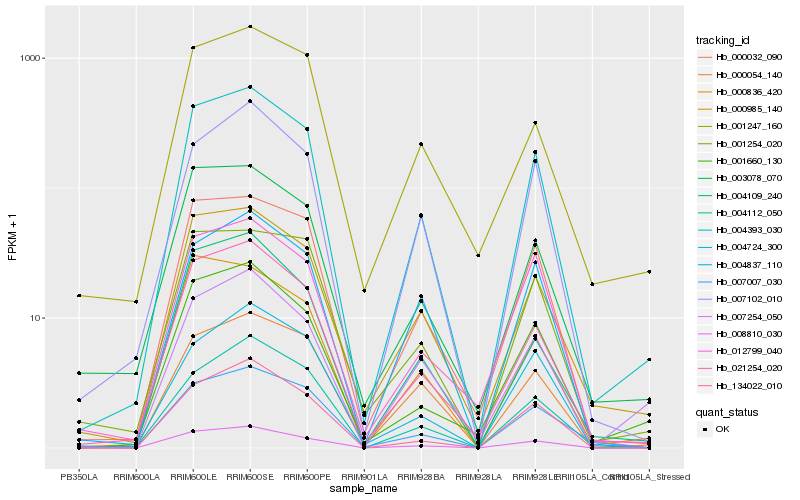
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004393_030 |
0.0 |
- |
- |
CBL-interacting protein kinase 6 [Populus trichocarpa] |
| 2 |
Hb_008810_030 |
0.0587286215 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_021254_020 |
0.0696730754 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 4 |
Hb_000032_090 |
0.077754265 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000985_140 |
0.0823760768 |
- |
- |
PREDICTED: uncharacterized protein LOC105643381 isoform X3 [Jatropha curcas] |
| 6 |
Hb_001660_130 |
0.0901624155 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR4 [Jatropha curcas] |
| 7 |
Hb_007007_030 |
0.0953978563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_007102_010 |
0.095469667 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 9 |
Hb_134022_010 |
0.0967389989 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 10 |
Hb_000054_140 |
0.0988335818 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1 [Jatropha curcas] |
| 11 |
Hb_004112_050 |
0.1032279241 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 12 |
Hb_004837_110 |
0.1035127365 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_012799_040 |
0.1061339215 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 14 |
Hb_003078_070 |
0.1075179978 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At3g25290 [Jatropha curcas] |
| 15 |
Hb_004109_240 |
0.1087639309 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 16 |
Hb_001254_020 |
0.1092400844 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 17 |
Hb_004724_300 |
0.111811129 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 18 |
Hb_000836_420 |
0.1146381613 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 19 |
Hb_007254_050 |
0.1164736713 |
- |
- |
PREDICTED: U-box domain-containing protein 28-like [Jatropha curcas] |
| 20 |
Hb_001247_160 |
0.1166962954 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |