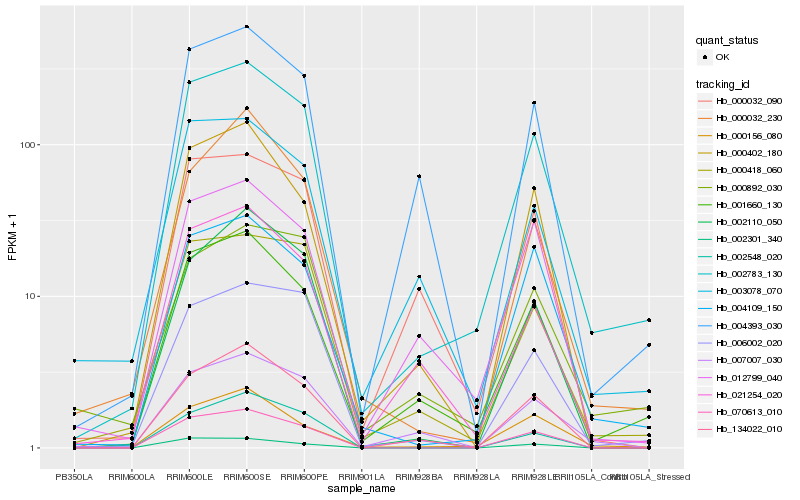
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002783_130 |
0.0 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_001660_130 |
0.0811385347 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR4 [Jatropha curcas] |
| 3 |
Hb_070613_010 |
0.106069545 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 4 |
Hb_000418_060 |
0.1123139897 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 5 |
Hb_004109_150 |
0.1140389172 |
- |
- |
glutathione-S-transferase tau 1 [Hevea brasiliensis] |
| 6 |
Hb_006002_020 |
0.1199776178 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 7 |
Hb_134022_010 |
0.1256758276 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 8 |
Hb_021254_020 |
0.1269751225 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 9 |
Hb_000402_180 |
0.130229752 |
- |
- |
PREDICTED: uncharacterized protein LOC105644975 [Jatropha curcas] |
| 10 |
Hb_004393_030 |
0.131697119 |
- |
- |
CBL-interacting protein kinase 6 [Populus trichocarpa] |
| 11 |
Hb_002110_050 |
0.1337735315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000156_080 |
0.1342396915 |
- |
- |
Potassium channel KAT2, putative [Ricinus communis] |
| 13 |
Hb_012799_040 |
0.1351558915 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 14 |
Hb_007007_030 |
0.1362216434 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002548_020 |
0.1368976441 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002301_340 |
0.1372952575 |
transcription factor |
TF Family: B3 |
leafy cotyledon 2 [Ricinus communis] |
| 17 |
Hb_003078_070 |
0.141475976 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At3g25290 [Jatropha curcas] |
| 18 |
Hb_000032_090 |
0.1423143992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000892_030 |
0.1431109803 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 20 |
Hb_000032_230 |
0.1444852884 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_20463 [Jatropha curcas] |