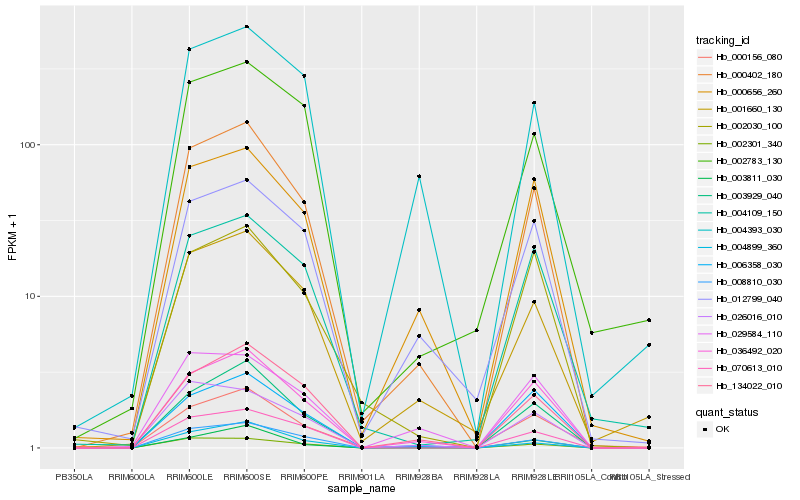
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000402_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644975 [Jatropha curcas] |
| 2 |
Hb_036492_020 |
0.0717797717 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 3 |
Hb_134022_010 |
0.0872620486 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 4 |
Hb_003929_040 |
0.0946581575 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_070613_010 |
0.1040731385 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 6 |
Hb_001660_130 |
0.1049783277 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR4 [Jatropha curcas] |
| 7 |
Hb_008810_030 |
0.1066543144 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_002301_340 |
0.1096684973 |
transcription factor |
TF Family: B3 |
leafy cotyledon 2 [Ricinus communis] |
| 9 |
Hb_000656_260 |
0.1118190744 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 10 |
Hb_026016_010 |
0.115562868 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 11 |
Hb_006358_030 |
0.1212621413 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 12 |
Hb_003811_030 |
0.1233024029 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 13 |
Hb_012799_040 |
0.1236850046 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 14 |
Hb_002030_100 |
0.1273138851 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 15 |
Hb_000156_080 |
0.1280001327 |
- |
- |
Potassium channel KAT2, putative [Ricinus communis] |
| 16 |
Hb_004393_030 |
0.128527868 |
- |
- |
CBL-interacting protein kinase 6 [Populus trichocarpa] |
| 17 |
Hb_004899_360 |
0.1299629608 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 18 |
Hb_002783_130 |
0.130229752 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_029584_110 |
0.1316237363 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 20 |
Hb_004109_150 |
0.1316968871 |
- |
- |
glutathione-S-transferase tau 1 [Hevea brasiliensis] |