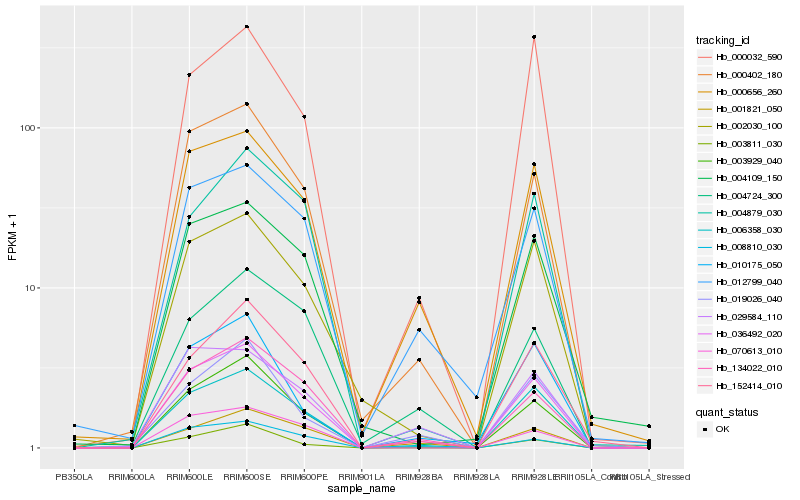
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_036492_020 |
0.0 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 2 |
Hb_006358_030 |
0.0683522387 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 3 |
Hb_000402_180 |
0.0717797717 |
- |
- |
PREDICTED: uncharacterized protein LOC105644975 [Jatropha curcas] |
| 4 |
Hb_000656_260 |
0.0793366725 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 5 |
Hb_134022_010 |
0.0903145575 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 6 |
Hb_003929_040 |
0.0985467058 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_152414_010 |
0.1085998905 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein PRUPE_ppa024232mg [Prunus persica] |
| 8 |
Hb_012799_040 |
0.108885878 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 9 |
Hb_002030_100 |
0.1116019006 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 10 |
Hb_003811_030 |
0.1177538644 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 11 |
Hb_001821_050 |
0.1193057945 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 12 |
Hb_029584_110 |
0.1194814455 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 13 |
Hb_000032_590 |
0.1229053889 |
- |
- |
PREDICTED: glutaredoxin domain-containing cysteine-rich protein 1 [Jatropha curcas] |
| 14 |
Hb_008810_030 |
0.1266669099 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_004109_150 |
0.1271327251 |
- |
- |
glutathione-S-transferase tau 1 [Hevea brasiliensis] |
| 16 |
Hb_070613_010 |
0.1292946974 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 17 |
Hb_004724_300 |
0.1302711809 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 18 |
Hb_019026_040 |
0.1304709837 |
- |
- |
S-locus lectin protein kinase family protein [Theobroma cacao] |
| 19 |
Hb_004879_030 |
0.1305047231 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 20 |
Hb_010175_050 |
0.1307177042 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |