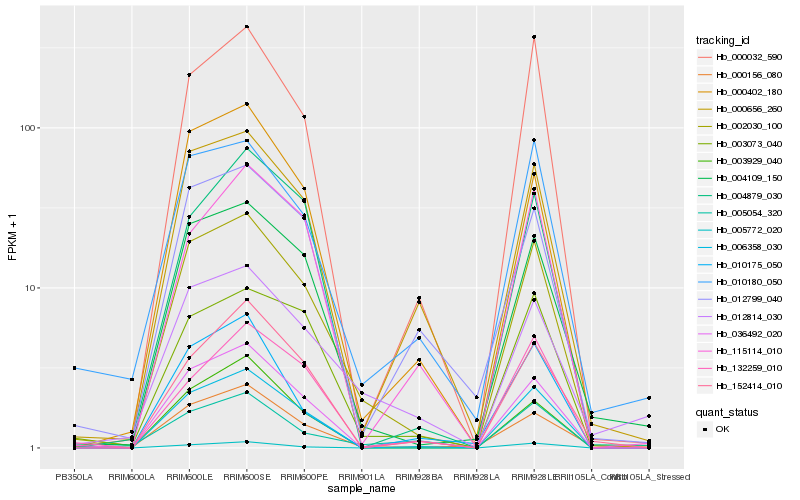
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002030_100 |
0.0 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 2 |
Hb_006358_030 |
0.0922467676 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 3 |
Hb_004109_150 |
0.0994324246 |
- |
- |
glutathione-S-transferase tau 1 [Hevea brasiliensis] |
| 4 |
Hb_036492_020 |
0.1116019006 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 5 |
Hb_000032_590 |
0.1219318636 |
- |
- |
PREDICTED: glutaredoxin domain-containing cysteine-rich protein 1 [Jatropha curcas] |
| 6 |
Hb_000656_260 |
0.1229638337 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 7 |
Hb_003073_040 |
0.1244010928 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.6-like isoform X2 [Populus euphratica] |
| 8 |
Hb_000402_180 |
0.1273138851 |
- |
- |
PREDICTED: uncharacterized protein LOC105644975 [Jatropha curcas] |
| 9 |
Hb_000156_080 |
0.1291759814 |
- |
- |
Potassium channel KAT2, putative [Ricinus communis] |
| 10 |
Hb_010180_050 |
0.1312976157 |
- |
- |
PREDICTED: CBL-interacting protein kinase 2 [Jatropha curcas] |
| 11 |
Hb_012814_030 |
0.1397632674 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 12 |
Hb_003929_040 |
0.1406502853 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_152414_010 |
0.1431001946 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein PRUPE_ppa024232mg [Prunus persica] |
| 14 |
Hb_005772_020 |
0.1451803019 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 15 |
Hb_012799_040 |
0.1462291777 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 16 |
Hb_005054_320 |
0.1479884443 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 17 |
Hb_004879_030 |
0.1488816875 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 18 |
Hb_132259_010 |
0.1568735553 |
- |
- |
PREDICTED: WAT1-related protein At5g64700 [Jatropha curcas] |
| 19 |
Hb_115114_010 |
0.1570169685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_010175_050 |
0.1617912862 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |