| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
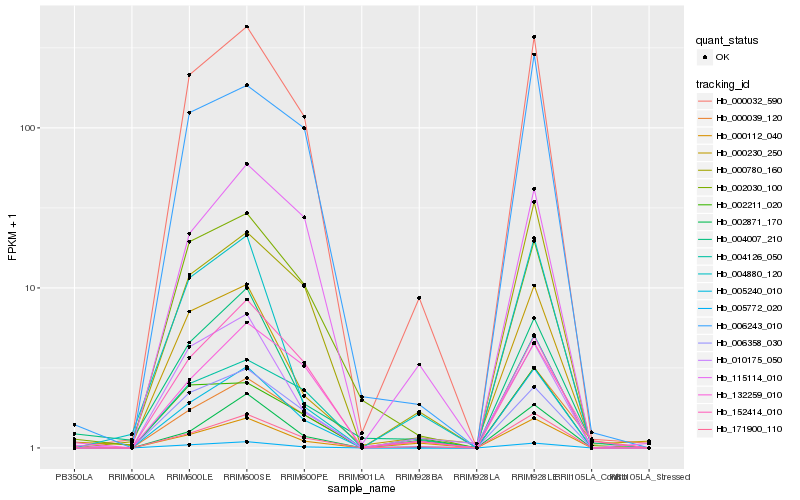
Hb_005240_010 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 2 |
Hb_005772_020 |
0.0684267574 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 3 |
Hb_000032_590 |
0.0787765865 |
- |
- |
PREDICTED: glutaredoxin domain-containing cysteine-rich protein 1 [Jatropha curcas] |
| 4 |
Hb_171900_110 |
0.0792683376 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT3-like [Populus euphratica] |
| 5 |
Hb_004126_050 |
0.1189347159 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VII.1 [Tarenaya hassleriana] |
| 6 |
Hb_000780_160 |
0.1262982246 |
- |
- |
hypothetical protein RCOM_0483300 [Ricinus communis] |
| 7 |
Hb_006358_030 |
0.1271595949 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 8 |
Hb_006243_010 |
0.1284317638 |
- |
- |
PREDICTED: cell wall protein IFF6 [Populus euphratica] |
| 9 |
Hb_000230_250 |
0.1457498521 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_132259_010 |
0.148320608 |
- |
- |
PREDICTED: WAT1-related protein At5g64700 [Jatropha curcas] |
| 11 |
Hb_000039_120 |
0.153678295 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |
| 12 |
Hb_000112_040 |
0.1594736048 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 13 |
Hb_010175_050 |
0.1612235779 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |
| 14 |
Hb_115114_010 |
0.1622385475 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002030_100 |
0.1656343649 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 16 |
Hb_002211_020 |
0.1696010081 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 17 |
Hb_152414_010 |
0.1703881299 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein PRUPE_ppa024232mg [Prunus persica] |
| 18 |
Hb_004880_120 |
0.1711388092 |
- |
- |
PREDICTED: calcium-binding protein PBP1 [Jatropha curcas] |
| 19 |
Hb_004007_210 |
0.1726956691 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 20 |
Hb_002871_170 |
0.172762122 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |