| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
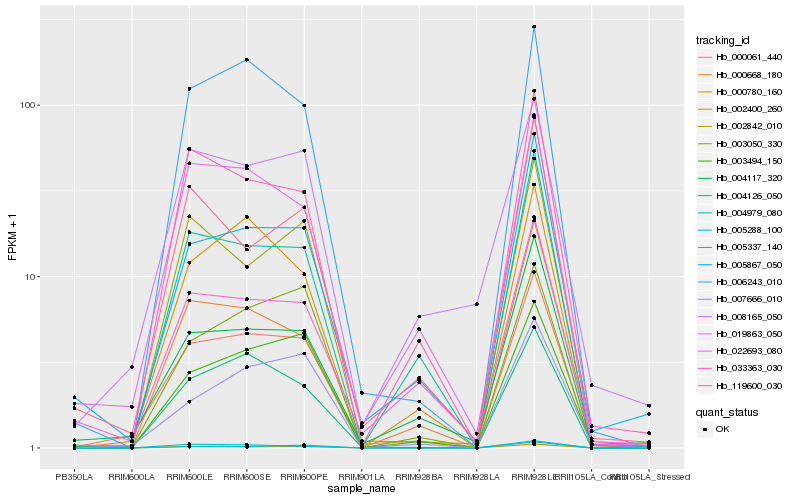
Hb_000061_440 |
0.0 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_004117_320 |
0.1119478411 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK5 [Jatropha curcas] |
| 3 |
Hb_005867_050 |
0.119938458 |
- |
- |
PREDICTED: protein ECERIFERUM 1 [Jatropha curcas] |
| 4 |
Hb_006243_010 |
0.1204376639 |
- |
- |
PREDICTED: cell wall protein IFF6 [Populus euphratica] |
| 5 |
Hb_004979_080 |
0.1222707334 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_019863_050 |
0.1240504429 |
- |
- |
PREDICTED: uncharacterized protein LOC103343426 [Prunus mume] |
| 7 |
Hb_004126_050 |
0.1252108709 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VII.1 [Tarenaya hassleriana] |
| 8 |
Hb_033363_030 |
0.1261641768 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002400_260 |
0.1264762297 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_000780_160 |
0.1271336404 |
- |
- |
hypothetical protein RCOM_0483300 [Ricinus communis] |
| 11 |
Hb_007666_010 |
0.1300889631 |
- |
- |
PREDICTED: uncharacterized protein LOC105649266 [Jatropha curcas] |
| 12 |
Hb_003494_150 |
0.1328386696 |
- |
- |
Silicon transporter, putative [Ricinus communis] |
| 13 |
Hb_000668_180 |
0.1331703172 |
- |
- |
hypothetical protein POPTR_0005s16490g [Populus trichocarpa] |
| 14 |
Hb_022693_080 |
0.1339492142 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 15 |
Hb_002842_010 |
0.1374763083 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 16 |
Hb_119600_030 |
0.1480198535 |
- |
- |
PREDICTED: uncharacterized protein LOC105635187 [Jatropha curcas] |
| 17 |
Hb_008165_050 |
0.1492330201 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 18 |
Hb_005337_140 |
0.1492960488 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: wall-associated receptor kinase 1-like [Jatropha curcas] |
| 19 |
Hb_005288_100 |
0.1495465104 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 20 |
Hb_003050_330 |
0.1507864178 |
- |
- |
hypothetical protein JCGZ_04591 [Jatropha curcas] |