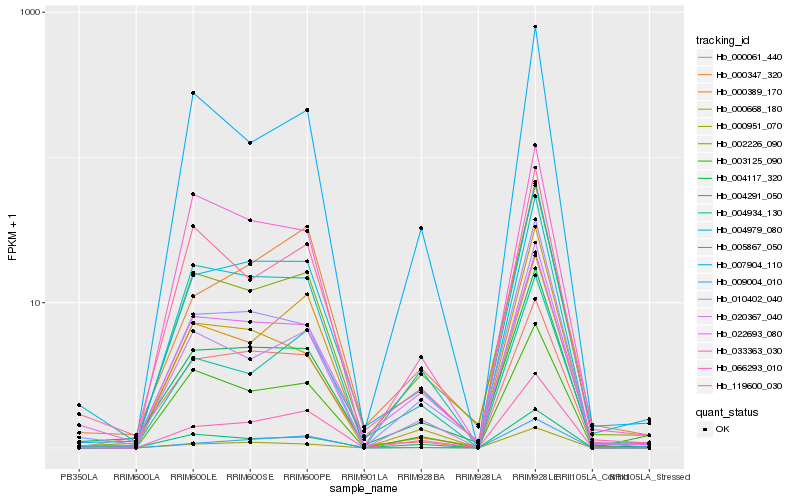
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005867_050 |
0.0 |
- |
- |
PREDICTED: protein ECERIFERUM 1 [Jatropha curcas] |
| 2 |
Hb_004117_320 |
0.0729655728 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK5 [Jatropha curcas] |
| 3 |
Hb_010402_040 |
0.0929627287 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 4 |
Hb_022693_080 |
0.0942016508 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 5 |
Hb_002226_090 |
0.1013188791 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 6 |
Hb_004979_080 |
0.1173989538 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_000061_440 |
0.119938458 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_000951_070 |
0.1227915291 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 9 |
Hb_066293_010 |
0.1294547906 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 10 |
Hb_000668_180 |
0.1302650493 |
- |
- |
hypothetical protein POPTR_0005s16490g [Populus trichocarpa] |
| 11 |
Hb_020367_040 |
0.1321524395 |
- |
- |
hypothetical protein JCGZ_01061 [Jatropha curcas] |
| 12 |
Hb_000347_320 |
0.1324000316 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 13 |
Hb_004934_130 |
0.1324240234 |
- |
- |
12-oxophytodienoate reductase 2 isoform 1 [Theobroma cacao] |
| 14 |
Hb_119600_030 |
0.1326758945 |
- |
- |
PREDICTED: uncharacterized protein LOC105635187 [Jatropha curcas] |
| 15 |
Hb_009004_010 |
0.1334243954 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 16 |
Hb_000389_170 |
0.1335314228 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 17 |
Hb_004291_050 |
0.1399036565 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 18 |
Hb_003125_090 |
0.1420484804 |
- |
- |
- |
| 19 |
Hb_033363_030 |
0.1421692135 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic [Jatropha curcas] |
| 20 |
Hb_007904_110 |
0.1458055471 |
- |
- |
PREDICTED: photosystem II core complex proteins psbY, chloroplastic-like [Populus euphratica] |