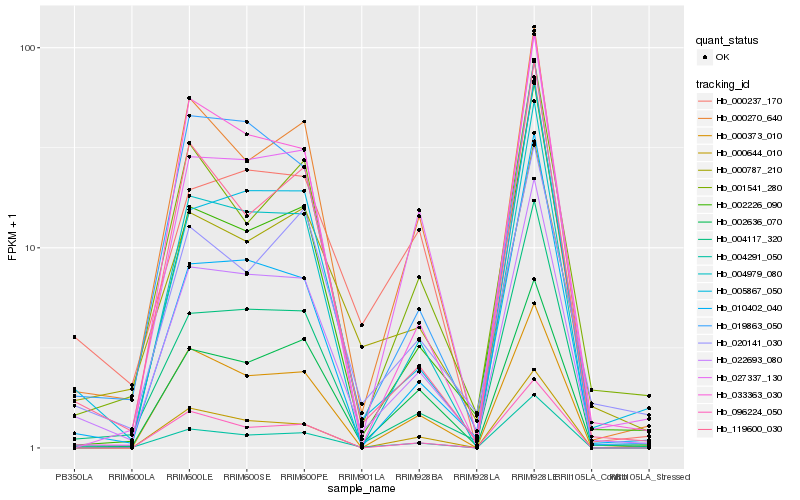
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022693_080 |
0.0 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 2 |
Hb_004117_320 |
0.087690673 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK5 [Jatropha curcas] |
| 3 |
Hb_005867_050 |
0.0942016508 |
- |
- |
PREDICTED: protein ECERIFERUM 1 [Jatropha curcas] |
| 4 |
Hb_000270_640 |
0.1011889158 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 5 |
Hb_004979_080 |
0.1042572769 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_004291_050 |
0.1124248174 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 7 |
Hb_019863_050 |
0.1132195314 |
- |
- |
PREDICTED: uncharacterized protein LOC103343426 [Prunus mume] |
| 8 |
Hb_002636_070 |
0.1136894851 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 9 |
Hb_002226_090 |
0.1188285136 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 10 |
Hb_010402_040 |
0.1206774667 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 11 |
Hb_027337_130 |
0.1207182016 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 12 |
Hb_001541_280 |
0.1226407476 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 13 |
Hb_020141_030 |
0.1234115211 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 14 |
Hb_033363_030 |
0.1236266807 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000237_170 |
0.1249702169 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 6 [Jatropha curcas] |
| 16 |
Hb_119600_030 |
0.1250775994 |
- |
- |
PREDICTED: uncharacterized protein LOC105635187 [Jatropha curcas] |
| 17 |
Hb_000644_010 |
0.1258654752 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000373_010 |
0.1286323132 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_000787_210 |
0.1301878952 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_096224_050 |
0.1302054394 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g31250 [Jatropha curcas] |