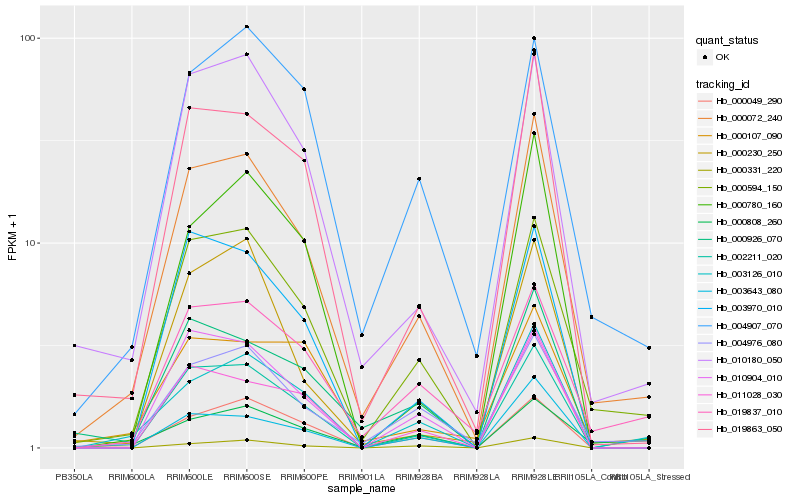
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000072_240 |
0.0 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_000594_150 |
0.0879462157 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic-like isoform X1 [Nelumbo nucifera] |
| 3 |
Hb_004976_080 |
0.1071276821 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 4 |
Hb_019863_050 |
0.1093780653 |
- |
- |
PREDICTED: uncharacterized protein LOC103343426 [Prunus mume] |
| 5 |
Hb_010180_050 |
0.114253723 |
- |
- |
PREDICTED: CBL-interacting protein kinase 2 [Jatropha curcas] |
| 6 |
Hb_003126_010 |
0.1239620468 |
- |
- |
PREDICTED: oligopeptide transporter 7-like isoform X1 [Malus domestica] |
| 7 |
Hb_002211_020 |
0.1273963457 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 8 |
Hb_019837_010 |
0.1343039354 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000926_070 |
0.1411093194 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004907_070 |
0.1464602084 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 11 |
Hb_000049_290 |
0.1481179275 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 12 |
Hb_011028_030 |
0.1506720577 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003970_010 |
0.1551534969 |
- |
- |
PREDICTED: sigma factor binding protein 1, chloroplastic-like [Populus euphratica] |
| 14 |
Hb_010904_010 |
0.1563455726 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 15 |
Hb_003643_080 |
0.1571211335 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 16 |
Hb_000331_220 |
0.1575306157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000780_160 |
0.1576204095 |
- |
- |
hypothetical protein RCOM_0483300 [Ricinus communis] |
| 18 |
Hb_000230_250 |
0.1583937231 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000808_260 |
0.1585357653 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 20 |
Hb_000107_090 |
0.1586920833 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |