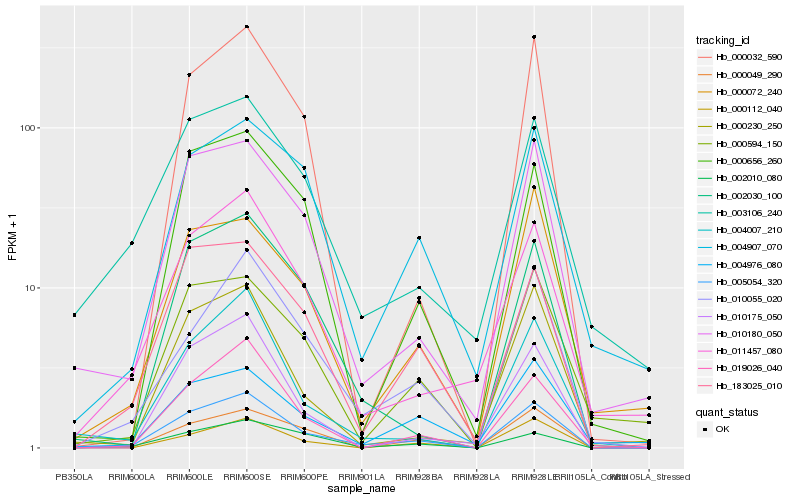
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005054_320 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 2 |
Hb_010180_050 |
0.1217642025 |
- |
- |
PREDICTED: CBL-interacting protein kinase 2 [Jatropha curcas] |
| 3 |
Hb_004007_210 |
0.1331420937 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 4 |
Hb_010055_020 |
0.1375470982 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 5 |
Hb_019026_040 |
0.1396079981 |
- |
- |
S-locus lectin protein kinase family protein [Theobroma cacao] |
| 6 |
Hb_183025_010 |
0.1404310319 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 7 |
Hb_000049_290 |
0.1433621859 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_002030_100 |
0.1479884443 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 9 |
Hb_000656_260 |
0.1501713074 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 10 |
Hb_000032_590 |
0.1505668382 |
- |
- |
PREDICTED: glutaredoxin domain-containing cysteine-rich protein 1 [Jatropha curcas] |
| 11 |
Hb_011457_080 |
0.1505778658 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000112_040 |
0.1511422317 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 13 |
Hb_000230_250 |
0.156258474 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_010175_050 |
0.1571855238 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |
| 15 |
Hb_003106_240 |
0.159328522 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 16 |
Hb_000594_150 |
0.1613840207 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic-like isoform X1 [Nelumbo nucifera] |
| 17 |
Hb_000072_240 |
0.1621591506 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_004907_070 |
0.1629303673 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 19 |
Hb_002010_080 |
0.1640190464 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 [Jatropha curcas] |
| 20 |
Hb_004976_080 |
0.1654680074 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |