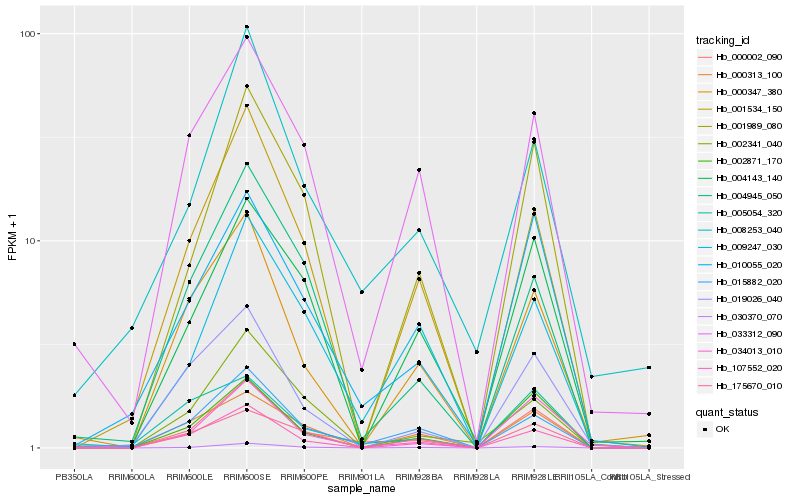
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000002_090 |
0.0 |
- |
- |
PREDICTED: RING-H2 finger protein ATL8-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_010055_020 |
0.1344692387 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 3 |
Hb_107552_020 |
0.1466551441 |
- |
- |
Ethylene-responsive transcription factor 1B, putative [Ricinus communis] |
| 4 |
Hb_002871_170 |
0.1480608509 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 5 |
Hb_034013_010 |
0.1532504583 |
- |
- |
hypothetical protein JCGZ_21944 [Jatropha curcas] |
| 6 |
Hb_001989_080 |
0.1565264827 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_030370_070 |
0.1651024098 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_015882_020 |
0.1657849589 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 9 |
Hb_001534_150 |
0.1701488539 |
- |
- |
PREDICTED: uncharacterized permease C29B12.14c [Jatropha curcas] |
| 10 |
Hb_008253_040 |
0.1769171594 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 11 |
Hb_009247_030 |
0.1796849069 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 12 |
Hb_019026_040 |
0.1803536605 |
- |
- |
S-locus lectin protein kinase family protein [Theobroma cacao] |
| 13 |
Hb_005054_320 |
0.182781709 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 14 |
Hb_000347_380 |
0.1831849013 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_004143_140 |
0.1835279255 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 16 |
Hb_000313_100 |
0.1850937063 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 17 |
Hb_175670_010 |
0.1871669649 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 18 |
Hb_004945_050 |
0.1877564422 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 19 |
Hb_033312_090 |
0.1878854128 |
- |
- |
PREDICTED: HMG-Y-related protein A-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_002341_040 |
0.188919359 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |