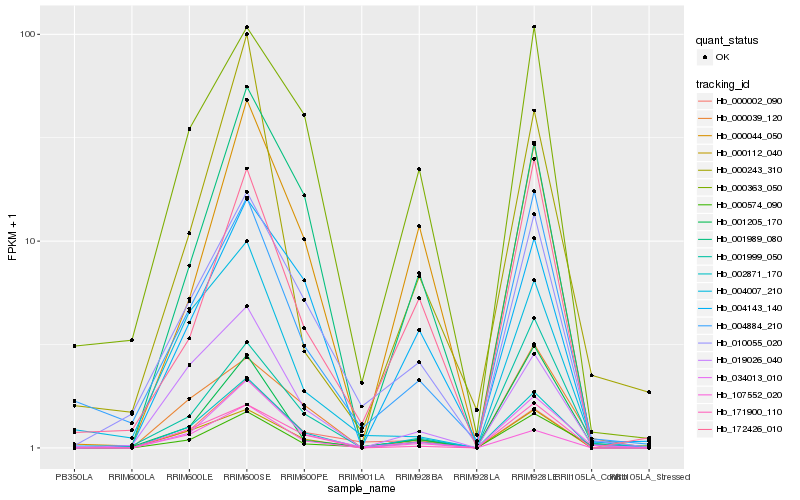
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034013_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_21944 [Jatropha curcas] |
| 2 |
Hb_002871_170 |
0.1201212892 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 3 |
Hb_004884_210 |
0.1372274375 |
- |
- |
PREDICTED: mitoferrin-like [Jatropha curcas] |
| 4 |
Hb_010055_020 |
0.1417479731 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 5 |
Hb_000002_090 |
0.1532504583 |
- |
- |
PREDICTED: RING-H2 finger protein ATL8-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_001989_080 |
0.1613984094 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_000112_040 |
0.1657733527 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 8 |
Hb_001205_170 |
0.1669021309 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 9 |
Hb_171900_110 |
0.1695238054 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT3-like [Populus euphratica] |
| 10 |
Hb_000574_090 |
0.1711758551 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 11 |
Hb_004007_210 |
0.1733804823 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000039_120 |
0.1746467828 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |
| 13 |
Hb_001999_050 |
0.1753871726 |
rubber biosynthesis |
Gene Name: CPT7 |
PREDICTED: dehydrodolichyl diphosphate synthase 2 [Jatropha curcas] |
| 14 |
Hb_172426_010 |
0.1766256846 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 15 |
Hb_000363_050 |
0.1792137287 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_019026_040 |
0.1808649736 |
- |
- |
S-locus lectin protein kinase family protein [Theobroma cacao] |
| 17 |
Hb_000243_310 |
0.1810608448 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 18 |
Hb_000044_050 |
0.1817662868 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_004143_140 |
0.1846675182 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 20 |
Hb_107552_020 |
0.1864047973 |
- |
- |
Ethylene-responsive transcription factor 1B, putative [Ricinus communis] |