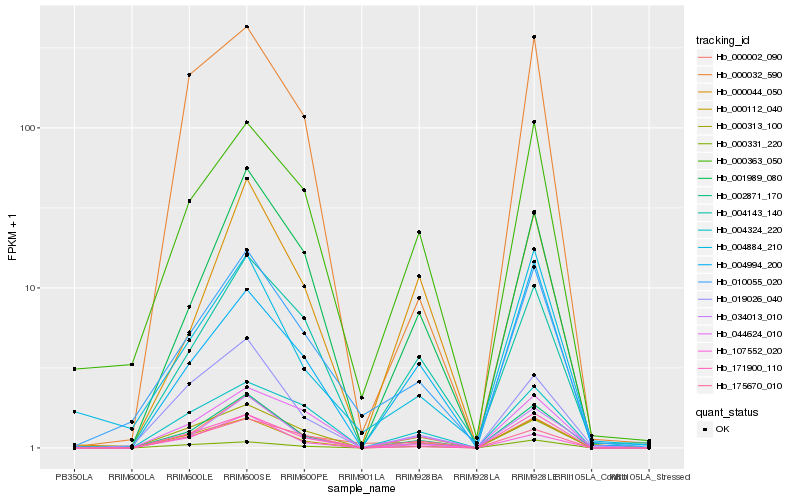
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002871_170 |
0.0 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 2 |
Hb_000112_040 |
0.0811956937 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 3 |
Hb_034013_010 |
0.1201212892 |
- |
- |
hypothetical protein JCGZ_21944 [Jatropha curcas] |
| 4 |
Hb_171900_110 |
0.124191903 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT3-like [Populus euphratica] |
| 5 |
Hb_001989_080 |
0.1303409803 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_107552_020 |
0.1363644881 |
- |
- |
Ethylene-responsive transcription factor 1B, putative [Ricinus communis] |
| 7 |
Hb_010055_020 |
0.1369983643 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 8 |
Hb_000331_220 |
0.1440045575 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000313_100 |
0.1475092565 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 10 |
Hb_000002_090 |
0.1480608509 |
- |
- |
PREDICTED: RING-H2 finger protein ATL8-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_019026_040 |
0.1488765229 |
- |
- |
S-locus lectin protein kinase family protein [Theobroma cacao] |
| 12 |
Hb_004143_140 |
0.1489267976 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 13 |
Hb_000044_050 |
0.1501856875 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000363_050 |
0.151976852 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_175670_010 |
0.1540938541 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 16 |
Hb_044624_010 |
0.1543390258 |
- |
- |
- |
| 17 |
Hb_000032_590 |
0.1594667728 |
- |
- |
PREDICTED: glutaredoxin domain-containing cysteine-rich protein 1 [Jatropha curcas] |
| 18 |
Hb_004884_210 |
0.1615977051 |
- |
- |
PREDICTED: mitoferrin-like [Jatropha curcas] |
| 19 |
Hb_004994_200 |
0.1630345412 |
- |
- |
PREDICTED: uncharacterized protein LOC105174340 [Sesamum indicum] |
| 20 |
Hb_004324_220 |
0.1636895339 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |