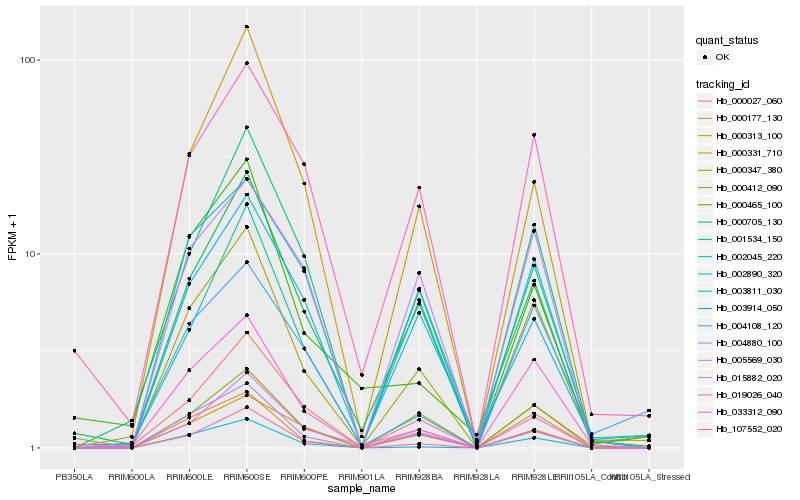
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_380 |
0.0 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_107552_020 |
0.0985833391 |
- |
- |
Ethylene-responsive transcription factor 1B, putative [Ricinus communis] |
| 3 |
Hb_000705_130 |
0.1250492266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_019026_040 |
0.1262108775 |
- |
- |
S-locus lectin protein kinase family protein [Theobroma cacao] |
| 5 |
Hb_001534_150 |
0.135706752 |
- |
- |
PREDICTED: uncharacterized permease C29B12.14c [Jatropha curcas] |
| 6 |
Hb_033312_090 |
0.1383349392 |
- |
- |
PREDICTED: HMG-Y-related protein A-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_004880_100 |
0.1414679982 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11300-like [Glycine max] |
| 8 |
Hb_015882_020 |
0.1456626098 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 9 |
Hb_003811_030 |
0.1464554876 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 10 |
Hb_000412_090 |
0.1492613925 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 11 |
Hb_002890_320 |
0.1497770261 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 12 |
Hb_000027_060 |
0.1506945285 |
- |
- |
hypothetical protein POPTR_0004s07470g [Populus trichocarpa] |
| 13 |
Hb_000177_130 |
0.1525777064 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 14 |
Hb_000313_100 |
0.154711955 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 15 |
Hb_000331_710 |
0.1583382696 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_005569_030 |
0.1642842042 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003914_050 |
0.1687663879 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 18 |
Hb_002045_220 |
0.1703283311 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004108_120 |
0.1706010312 |
- |
- |
PREDICTED: EID1-like F-box protein 3 [Jatropha curcas] |
| 20 |
Hb_000465_100 |
0.1710485006 |
- |
- |
catalytic, putative [Ricinus communis] |