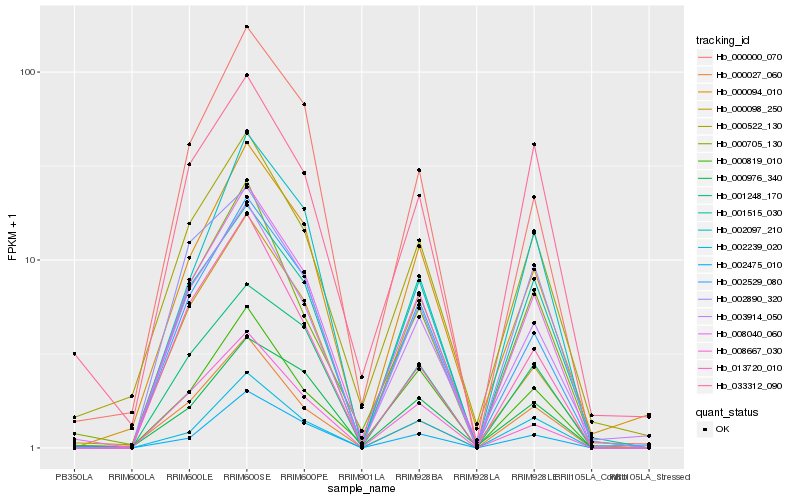
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008040_060 |
0.0 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 2 |
Hb_002529_080 |
0.0783161902 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase [Jatropha curcas] |
| 3 |
Hb_001248_170 |
0.0891769134 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 4 |
Hb_003914_050 |
0.0955264443 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 5 |
Hb_000027_060 |
0.101406993 |
- |
- |
hypothetical protein POPTR_0004s07470g [Populus trichocarpa] |
| 6 |
Hb_000000_070 |
0.1018783509 |
- |
- |
allene oxide synthase [Hevea brasiliensis] |
| 7 |
Hb_008667_030 |
0.101881759 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 8 |
Hb_001515_030 |
0.102017168 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000094_010 |
0.1027238247 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 10 |
Hb_000819_010 |
0.1027511541 |
- |
- |
Receptor-like protein kinase 1, putative [Theobroma cacao] |
| 11 |
Hb_000705_130 |
0.1035818322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000522_130 |
0.103896561 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 13 |
Hb_002239_020 |
0.1109647966 |
- |
- |
MLO, putative [Theobroma cacao] |
| 14 |
Hb_000976_340 |
0.1130851507 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_002097_210 |
0.1175578499 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000098_250 |
0.1196518335 |
- |
- |
UDP-glucose flavonoid 3-O-glucosyltransferase [Medicago truncatula] |
| 17 |
Hb_033312_090 |
0.1200517697 |
- |
- |
PREDICTED: HMG-Y-related protein A-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_002475_010 |
0.1200603698 |
- |
- |
hypothetical protein JCGZ_22530 [Jatropha curcas] |
| 19 |
Hb_002890_320 |
0.1211681448 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 20 |
Hb_013720_010 |
0.121555676 |
- |
- |
Putative DNA repair RAD23-3 -like protein [Gossypium arboreum] |