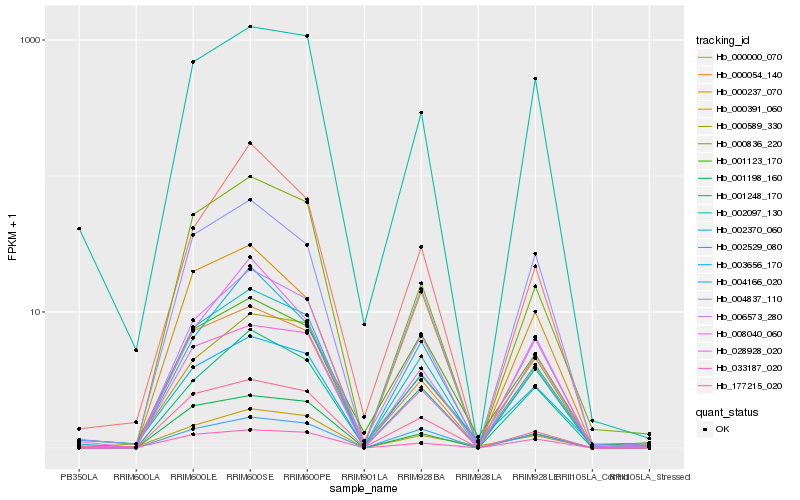
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002370_060 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Populus euphratica] |
| 2 |
Hb_000054_140 |
0.0737490576 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1 [Jatropha curcas] |
| 3 |
Hb_177215_020 |
0.0870348401 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 4 |
Hb_000836_220 |
0.0874146827 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_001248_170 |
0.0915834466 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 6 |
Hb_006573_280 |
0.0936454349 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 7 |
Hb_001198_160 |
0.1070761778 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 8 |
Hb_002097_130 |
0.1189412327 |
- |
- |
tonoplast intrinsic protein [Hevea brasiliensis] |
| 9 |
Hb_004837_110 |
0.12088534 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004166_020 |
0.1211610468 |
- |
- |
- |
| 11 |
Hb_002529_080 |
0.1223527652 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase [Jatropha curcas] |
| 12 |
Hb_003656_170 |
0.124286701 |
- |
- |
PREDICTED: phenylalanine ammonia-lyase-like [Jatropha curcas] |
| 13 |
Hb_000589_330 |
0.1274671073 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_000391_060 |
0.1301851596 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 15 |
Hb_000000_070 |
0.131191455 |
- |
- |
allene oxide synthase [Hevea brasiliensis] |
| 16 |
Hb_001123_170 |
0.1329892699 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_033187_020 |
0.1338086185 |
- |
- |
hypothetical protein CARUB_v10004250mg, partial [Capsella rubella] |
| 18 |
Hb_000237_070 |
0.1347180803 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 19 |
Hb_008040_060 |
0.1356384729 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 20 |
Hb_028928_020 |
0.1363220992 |
- |
- |
hypothetical protein CICLE_v10018504mg [Citrus clementina] |