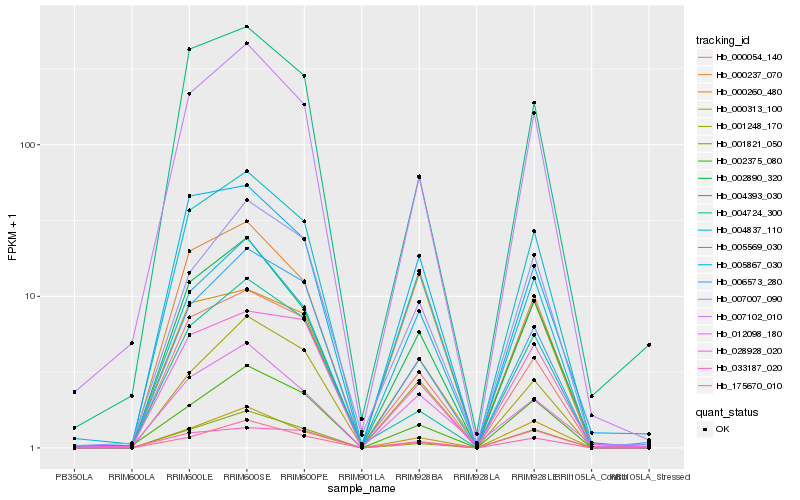
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004837_110 |
0.0 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007102_010 |
0.0615833644 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 3 |
Hb_002890_320 |
0.0657093002 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 4 |
Hb_000054_140 |
0.0856727363 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1 [Jatropha curcas] |
| 5 |
Hb_001821_050 |
0.0946597001 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 6 |
Hb_028928_020 |
0.1011841816 |
- |
- |
hypothetical protein CICLE_v10018504mg [Citrus clementina] |
| 7 |
Hb_002375_080 |
0.1014586581 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_007007_090 |
0.1028117263 |
- |
- |
phospholipase d beta, putative [Ricinus communis] |
| 9 |
Hb_004393_030 |
0.1035127365 |
- |
- |
CBL-interacting protein kinase 6 [Populus trichocarpa] |
| 10 |
Hb_006573_280 |
0.1056241914 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 11 |
Hb_005569_030 |
0.1062914026 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 isoform X1 [Jatropha curcas] |
| 12 |
Hb_033187_020 |
0.1102091599 |
- |
- |
hypothetical protein CARUB_v10004250mg, partial [Capsella rubella] |
| 13 |
Hb_001248_170 |
0.1119364486 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 14 |
Hb_000313_100 |
0.1128611451 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 15 |
Hb_000260_480 |
0.1142222547 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 16 |
Hb_004724_300 |
0.1183685725 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 17 |
Hb_000237_070 |
0.1184094182 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 18 |
Hb_012098_180 |
0.1189812175 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 19 |
Hb_175670_010 |
0.1193015494 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 20 |
Hb_005867_030 |
0.1195671777 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |