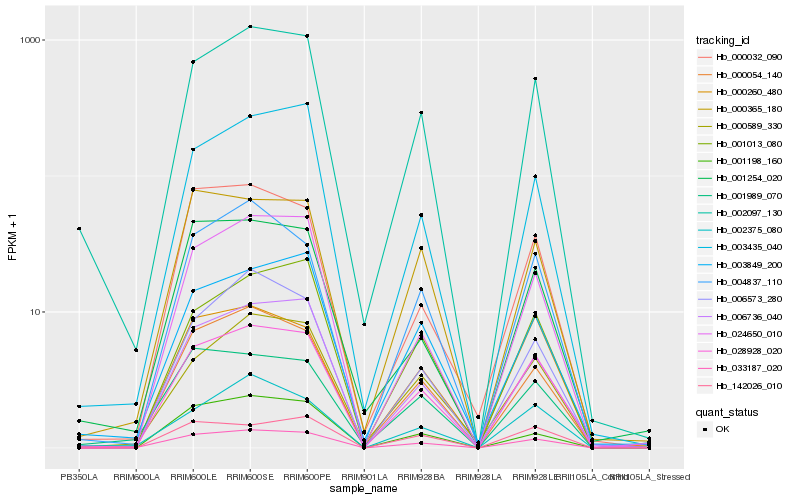
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033187_020 |
0.0 |
- |
- |
hypothetical protein CARUB_v10004250mg, partial [Capsella rubella] |
| 2 |
Hb_028928_020 |
0.0675148881 |
- |
- |
hypothetical protein CICLE_v10018504mg [Citrus clementina] |
| 3 |
Hb_000054_140 |
0.0937474022 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1 [Jatropha curcas] |
| 4 |
Hb_002097_130 |
0.0975898202 |
- |
- |
tonoplast intrinsic protein [Hevea brasiliensis] |
| 5 |
Hb_006736_040 |
0.1002094503 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase, basic isoform-like [Populus euphratica] |
| 6 |
Hb_000365_180 |
0.1047508933 |
- |
- |
PREDICTED: protein LURP-one-related 8-like [Pyrus x bretschneideri] |
| 7 |
Hb_024650_010 |
0.1049797789 |
- |
- |
PREDICTED: BAHD acyltransferase DCR [Jatropha curcas] |
| 8 |
Hb_004837_110 |
0.1102091599 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001989_070 |
0.1154034247 |
- |
- |
PREDICTED: uncharacterized calcium-binding protein At1g02270 [Jatropha curcas] |
| 10 |
Hb_000032_090 |
0.1190502678 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000589_330 |
0.1192709916 |
- |
- |
kinase, putative [Ricinus communis] |
| 12 |
Hb_001198_160 |
0.1207125653 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 13 |
Hb_003435_040 |
0.120721517 |
- |
- |
tonoplast intrinsic protein [Hevea brasiliensis] |
| 14 |
Hb_006573_280 |
0.1214435416 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 15 |
Hb_003849_200 |
0.1231719165 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 16 |
Hb_001254_020 |
0.1241606687 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 17 |
Hb_000260_480 |
0.1256912074 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 18 |
Hb_001013_080 |
0.126990985 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 19 |
Hb_142026_010 |
0.1270292881 |
- |
- |
PREDICTED: probable mannitol dehydrogenase [Jatropha curcas] |
| 20 |
Hb_002375_080 |
0.1280260902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |