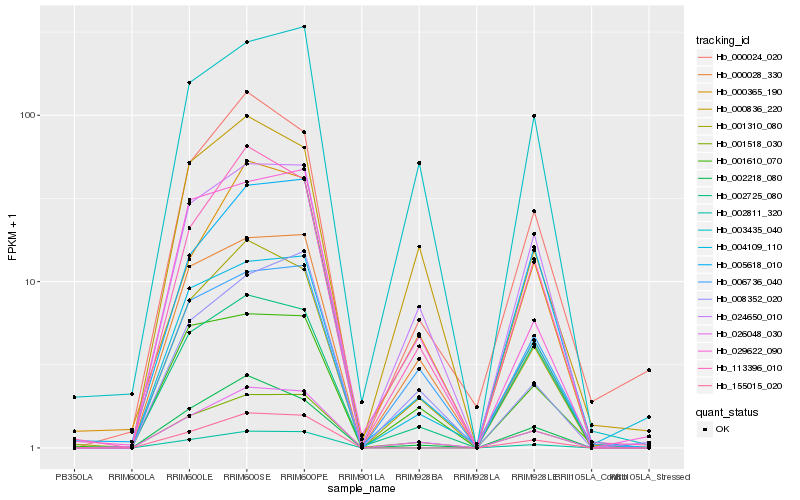
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026048_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001518_030 |
0.050585486 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 3 |
Hb_001310_080 |
0.0675242807 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein AZF2-like [Jatropha curcas] |
| 4 |
Hb_113396_010 |
0.0791389563 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_000028_330 |
0.084648359 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 9-like [Pyrus x bretschneideri] |
| 6 |
Hb_024650_010 |
0.0869499033 |
- |
- |
PREDICTED: BAHD acyltransferase DCR [Jatropha curcas] |
| 7 |
Hb_000365_190 |
0.0985460277 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 8 |
Hb_002218_080 |
0.1055265496 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 9 |
Hb_008352_020 |
0.1078839557 |
- |
- |
hypothetical protein [Cecembia lonarensis] |
| 10 |
Hb_003435_040 |
0.1095409749 |
- |
- |
tonoplast intrinsic protein [Hevea brasiliensis] |
| 11 |
Hb_006736_040 |
0.1100406981 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase, basic isoform-like [Populus euphratica] |
| 12 |
Hb_155015_020 |
0.1118393634 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 13 |
Hb_002811_320 |
0.1137539353 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 14 |
Hb_005618_010 |
0.1164970614 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0014s10680g [Populus trichocarpa] |
| 15 |
Hb_004109_110 |
0.1181930091 |
- |
- |
dioxygenase family protein [Populus trichocarpa] |
| 16 |
Hb_000024_020 |
0.1184088818 |
- |
- |
PREDICTED: uncharacterized protein LOC105636896 [Jatropha curcas] |
| 17 |
Hb_002725_080 |
0.1245901936 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 18 |
Hb_029622_090 |
0.1257356746 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 2-like [Jatropha curcas] |
| 19 |
Hb_001610_070 |
0.1258054511 |
- |
- |
hypothetical protein MIMGU_mgv1a022867mg [Erythranthe guttata] |
| 20 |
Hb_000836_220 |
0.1258343804 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |