| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
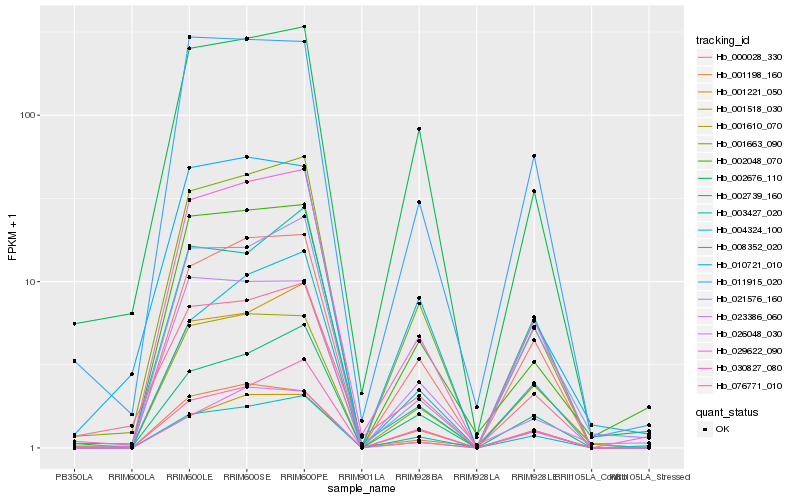
Hb_029622_090 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 2-like [Jatropha curcas] |
| 2 |
Hb_001663_090 |
0.0594302209 |
- |
- |
ribonuclease t2, putative [Ricinus communis] |
| 3 |
Hb_000028_330 |
0.082772954 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 9-like [Pyrus x bretschneideri] |
| 4 |
Hb_002048_070 |
0.0930364635 |
- |
- |
hypothetical protein JCGZ_15146 [Jatropha curcas] |
| 5 |
Hb_011915_020 |
0.0985404928 |
- |
- |
PREDICTED: arogenate dehydratase/prephenate dehydratase 6, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_021576_160 |
0.1016605819 |
transcription factor |
TF Family: WRKY |
putative WRKY transcription factor family protein [Populus trichocarpa] |
| 7 |
Hb_004324_100 |
0.1085702211 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 8 |
Hb_001610_070 |
0.1104018744 |
- |
- |
hypothetical protein MIMGU_mgv1a022867mg [Erythranthe guttata] |
| 9 |
Hb_001518_030 |
0.1114621133 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 10 |
Hb_076771_010 |
0.1121924621 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 11 |
Hb_008352_020 |
0.1124680852 |
- |
- |
hypothetical protein [Cecembia lonarensis] |
| 12 |
Hb_010721_010 |
0.1127994378 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Populus euphratica] |
| 13 |
Hb_002739_160 |
0.1130272111 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_002676_110 |
0.1172064359 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 2-like [Gossypium raimondii] |
| 15 |
Hb_001198_160 |
0.120036828 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 16 |
Hb_030827_080 |
0.1200978197 |
- |
- |
PREDICTED: uncharacterized protein LOC105648491 [Jatropha curcas] |
| 17 |
Hb_001221_050 |
0.1211747428 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |
| 18 |
Hb_023386_060 |
0.1213073984 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 19 |
Hb_026048_030 |
0.1257356746 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003427_020 |
0.1263613301 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |