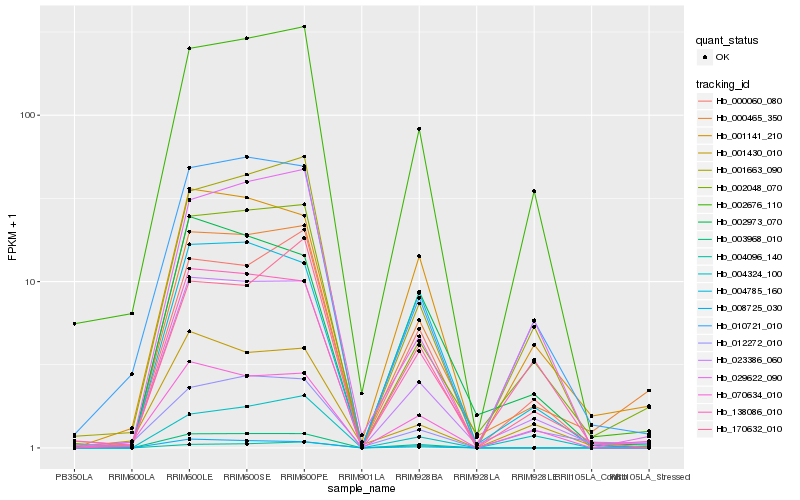
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000465_350 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 2 [Jatropha curcas] |
| 2 |
Hb_002048_070 |
0.092465411 |
- |
- |
hypothetical protein JCGZ_15146 [Jatropha curcas] |
| 3 |
Hb_023386_060 |
0.0943051332 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 4 |
Hb_001141_210 |
0.135679862 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 5 |
Hb_002676_110 |
0.138631099 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 2-like [Gossypium raimondii] |
| 6 |
Hb_000060_080 |
0.1386624091 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 2 protein, putative [Ricinus communis] |
| 7 |
Hb_003968_010 |
0.1430435265 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_010721_010 |
0.1434022838 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Populus euphratica] |
| 9 |
Hb_001663_090 |
0.1532766169 |
- |
- |
ribonuclease t2, putative [Ricinus communis] |
| 10 |
Hb_138086_010 |
0.155440478 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 11 |
Hb_170632_010 |
0.1557761207 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 12 |
Hb_004785_160 |
0.1599565478 |
- |
- |
PREDICTED: U-box domain-containing protein 28-like [Jatropha curcas] |
| 13 |
Hb_004096_140 |
0.1603529112 |
- |
- |
Lipase, putative [Ricinus communis] |
| 14 |
Hb_008725_030 |
0.1604338579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002973_070 |
0.1607958931 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 16 |
Hb_029622_090 |
0.1625948909 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 2-like [Jatropha curcas] |
| 17 |
Hb_012272_010 |
0.1629528453 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 18 |
Hb_004324_100 |
0.1631725595 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 19 |
Hb_001430_010 |
0.1645802448 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_070634_010 |
0.1678693362 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |