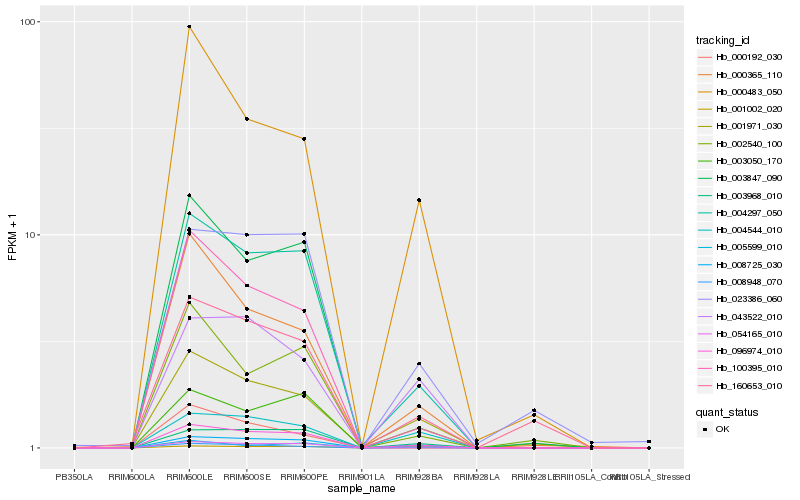
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_096974_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105123384 isoform X1 [Populus euphratica] |
| 2 |
Hb_004297_050 |
0.0368614837 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_008725_030 |
0.0832535534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003968_010 |
0.1062542134 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_008948_070 |
0.1088659446 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_100395_010 |
0.1194103547 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_001971_030 |
0.1307266686 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 8 |
Hb_003050_170 |
0.136574115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_023386_060 |
0.1381357228 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 10 |
Hb_160653_010 |
0.1438482897 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 11 |
Hb_000365_110 |
0.1452363176 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 12 |
Hb_005599_010 |
0.1459463016 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 13 |
Hb_054165_010 |
0.1512973843 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 14 |
Hb_001002_020 |
0.1541879453 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_043522_010 |
0.1549920035 |
- |
- |
- |
| 16 |
Hb_000483_050 |
0.1561803735 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 17 |
Hb_000192_030 |
0.1567708752 |
- |
- |
PREDICTED: TATA-box-binding protein 2 isoform X1 [Setaria italica] |
| 18 |
Hb_004544_010 |
0.1572391694 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Jatropha curcas] |
| 19 |
Hb_002540_100 |
0.1575367516 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 20 |
Hb_003847_090 |
0.1579974509 |
- |
- |
PREDICTED: tyrosine decarboxylase 1-like [Jatropha curcas] |