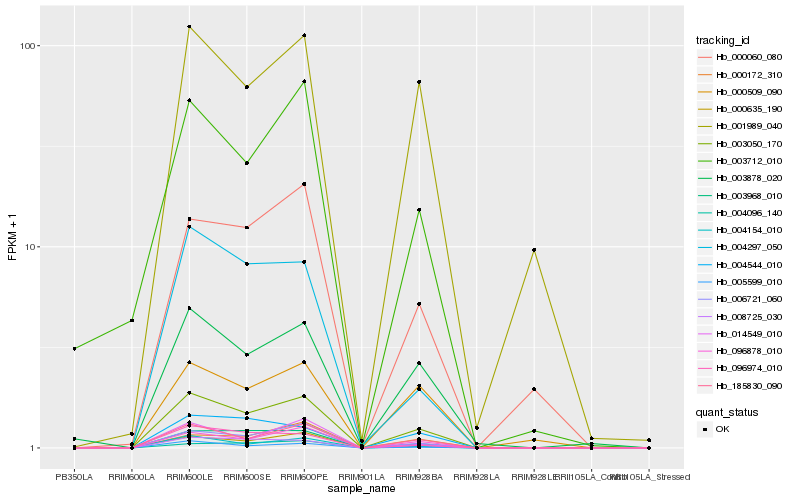
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003050_170 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000635_190 |
0.077620745 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103322127 [Prunus mume] |
| 3 |
Hb_185830_090 |
0.0823907119 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_096878_010 |
0.0887538785 |
- |
- |
hypothetical protein POPTR_0112s002001g, partial [Populus trichocarpa] |
| 5 |
Hb_006721_060 |
0.0962666277 |
- |
- |
RnaseH (mitochondrion) [Malus domestica] |
| 6 |
Hb_008725_030 |
0.1063951284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003968_010 |
0.1138478648 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_004154_010 |
0.1174988822 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 9 |
Hb_003878_020 |
0.1254205184 |
- |
- |
Epidermis-specific secreted glycoprotein EP1 precursor, putative [Ricinus communis] |
| 10 |
Hb_005599_010 |
0.1266865396 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 11 |
Hb_096974_010 |
0.136574115 |
- |
- |
PREDICTED: uncharacterized protein LOC105123384 isoform X1 [Populus euphratica] |
| 12 |
Hb_004096_140 |
0.13883143 |
- |
- |
Lipase, putative [Ricinus communis] |
| 13 |
Hb_004297_050 |
0.1396747141 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_000060_080 |
0.1410443968 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 2 protein, putative [Ricinus communis] |
| 15 |
Hb_014549_010 |
0.1465616151 |
- |
- |
PREDICTED: uncharacterized protein LOC101512634 [Cicer arietinum] |
| 16 |
Hb_000509_090 |
0.1491649713 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 17 |
Hb_001989_040 |
0.1504431615 |
- |
- |
PREDICTED: neurofilament heavy polypeptide [Jatropha curcas] |
| 18 |
Hb_004544_010 |
0.1520312135 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Jatropha curcas] |
| 19 |
Hb_003712_010 |
0.1520615394 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_000172_310 |
0.1550308841 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 21 [Jatropha curcas] |