| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
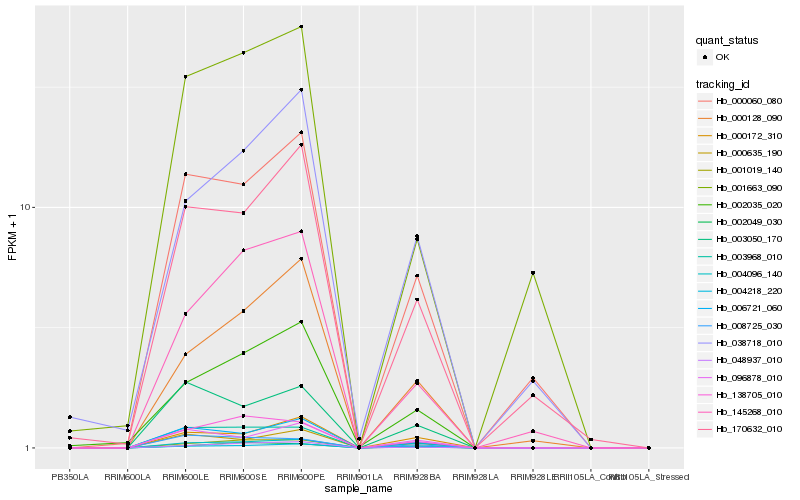
Hb_004096_140 |
0.0 |
- |
- |
Lipase, putative [Ricinus communis] |
| 2 |
Hb_001019_140 |
0.0827505144 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_006721_060 |
0.0889269369 |
- |
- |
RnaseH (mitochondrion) [Malus domestica] |
| 4 |
Hb_048937_010 |
0.09123738 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1 [Nelumbo nucifera] |
| 5 |
Hb_003968_010 |
0.0935872809 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_138705_010 |
0.1125194521 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 7 |
Hb_000060_080 |
0.1136615406 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 2 protein, putative [Ricinus communis] |
| 8 |
Hb_000635_190 |
0.1137348105 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103322127 [Prunus mume] |
| 9 |
Hb_002035_020 |
0.1167915128 |
transcription factor |
TF Family: TRAF |
hypothetical protein B456_011G050200 [Gossypium raimondii] |
| 10 |
Hb_145268_010 |
0.1188013659 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Jatropha curcas] |
| 11 |
Hb_170632_010 |
0.1222779731 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 12 |
Hb_000172_310 |
0.1260530336 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 21 [Jatropha curcas] |
| 13 |
Hb_000128_090 |
0.1268032059 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 14 |
Hb_096878_010 |
0.1386579888 |
- |
- |
hypothetical protein POPTR_0112s002001g, partial [Populus trichocarpa] |
| 15 |
Hb_003050_170 |
0.13883143 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001663_090 |
0.1392570745 |
- |
- |
ribonuclease t2, putative [Ricinus communis] |
| 17 |
Hb_004218_220 |
0.1398923658 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 18 |
Hb_002049_030 |
0.1441487277 |
- |
- |
PREDICTED: uncharacterized protein LOC105635940 isoform X3 [Jatropha curcas] |
| 19 |
Hb_038718_010 |
0.1504792763 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 20 |
Hb_008725_030 |
0.1555258337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |