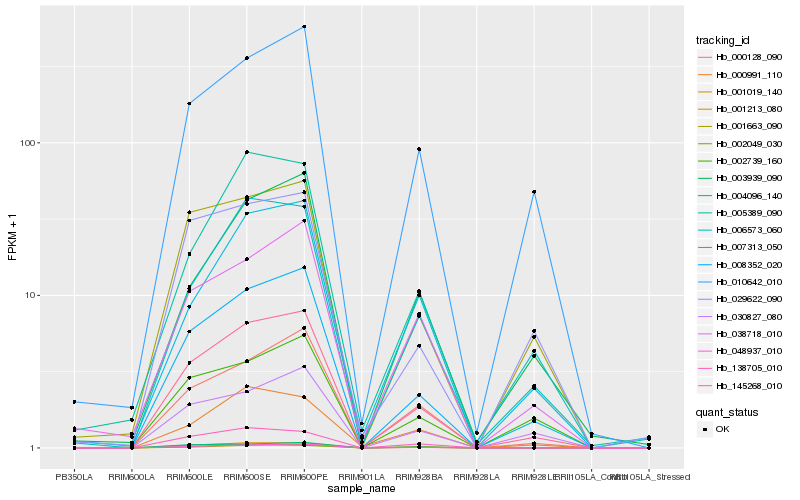
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_145268_010 |
0.0 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Jatropha curcas] |
| 2 |
Hb_048937_010 |
0.0739369807 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1 [Nelumbo nucifera] |
| 3 |
Hb_000128_090 |
0.0998941162 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 4 |
Hb_008352_020 |
0.100636512 |
- |
- |
hypothetical protein [Cecembia lonarensis] |
| 5 |
Hb_010642_010 |
0.1037432361 |
- |
- |
PREDICTED: aquaporin TIP2-1 [Jatropha curcas] |
| 6 |
Hb_001019_140 |
0.1067644599 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_001663_090 |
0.1118790325 |
- |
- |
ribonuclease t2, putative [Ricinus communis] |
| 8 |
Hb_004096_140 |
0.1188013659 |
- |
- |
Lipase, putative [Ricinus communis] |
| 9 |
Hb_030827_080 |
0.1228022767 |
- |
- |
PREDICTED: uncharacterized protein LOC105648491 [Jatropha curcas] |
| 10 |
Hb_001213_080 |
0.1230077128 |
- |
- |
PREDICTED: tubulin beta chain-like [Jatropha curcas] |
| 11 |
Hb_006573_060 |
0.1259342686 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 12 |
Hb_002049_030 |
0.1276748946 |
- |
- |
PREDICTED: uncharacterized protein LOC105635940 isoform X3 [Jatropha curcas] |
| 13 |
Hb_138705_010 |
0.1282198554 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 14 |
Hb_005389_090 |
0.1290779248 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA27, putative [Ricinus communis] |
| 15 |
Hb_003939_090 |
0.1297211841 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_038718_010 |
0.1303615662 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 17 |
Hb_007313_050 |
0.1315939756 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 18 |
Hb_002739_160 |
0.1322294729 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_000991_110 |
0.134185346 |
- |
- |
PREDICTED: uncharacterized protein LOC105636336 [Jatropha curcas] |
| 20 |
Hb_029622_090 |
0.1355766831 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 2-like [Jatropha curcas] |