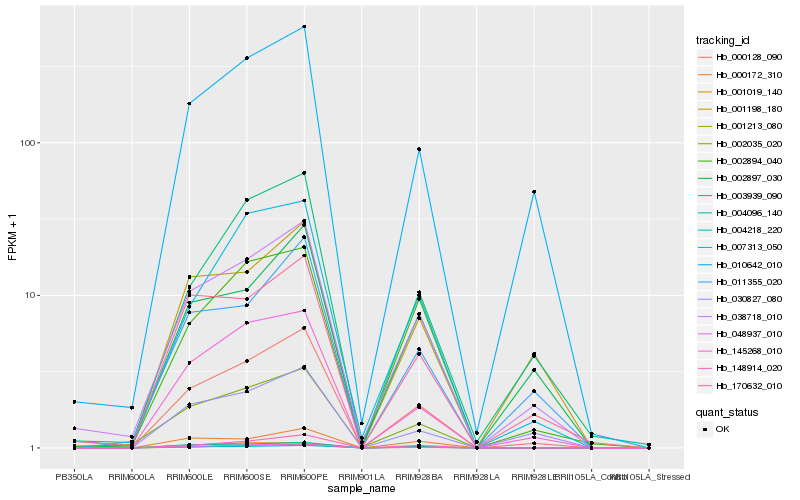
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000128_090 |
0.0 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 2 |
Hb_038718_010 |
0.0819056886 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 3 |
Hb_148914_020 |
0.0851725329 |
- |
- |
PREDICTED: uncharacterized protein LOC105637821 [Jatropha curcas] |
| 4 |
Hb_145268_010 |
0.0998941162 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Jatropha curcas] |
| 5 |
Hb_001213_080 |
0.104055819 |
- |
- |
PREDICTED: tubulin beta chain-like [Jatropha curcas] |
| 6 |
Hb_010642_010 |
0.1042088967 |
- |
- |
PREDICTED: aquaporin TIP2-1 [Jatropha curcas] |
| 7 |
Hb_048937_010 |
0.1093143325 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1 [Nelumbo nucifera] |
| 8 |
Hb_002035_020 |
0.1165227703 |
transcription factor |
TF Family: TRAF |
hypothetical protein B456_011G050200 [Gossypium raimondii] |
| 9 |
Hb_003939_090 |
0.1192404722 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_011355_020 |
0.1207983223 |
- |
- |
extensin, proline-rich protein, putative [Ricinus communis] |
| 11 |
Hb_007313_050 |
0.1215584825 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 12 |
Hb_170632_010 |
0.1247793509 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 13 |
Hb_004096_140 |
0.1268032059 |
- |
- |
Lipase, putative [Ricinus communis] |
| 14 |
Hb_030827_080 |
0.1302102094 |
- |
- |
PREDICTED: uncharacterized protein LOC105648491 [Jatropha curcas] |
| 15 |
Hb_002897_030 |
0.133095029 |
- |
- |
PREDICTED: methylesterase 17 [Jatropha curcas] |
| 16 |
Hb_000172_310 |
0.1338891712 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 21 [Jatropha curcas] |
| 17 |
Hb_002894_040 |
0.1340449036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004218_220 |
0.135343193 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_001198_180 |
0.1365432299 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 20 |
Hb_001019_140 |
0.1381244444 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |