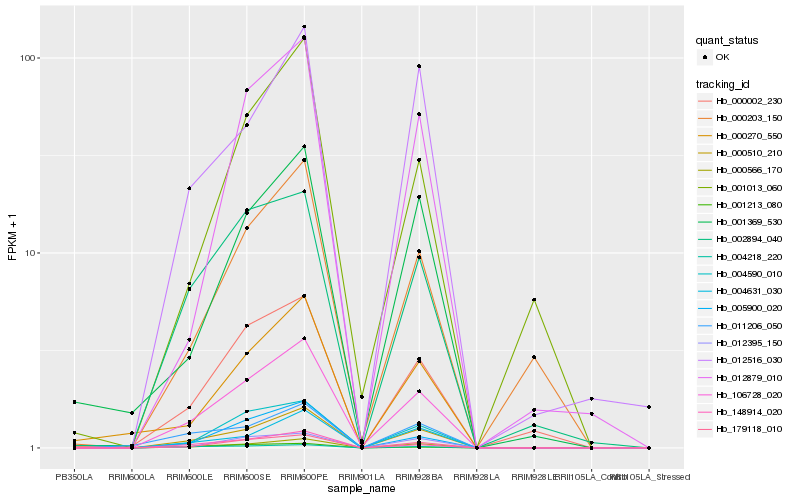
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_106728_020 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_000510_210 |
0.0605507694 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_000566_170 |
0.0688180363 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL9 homolog isoform X4 [Jatropha curcas] |
| 4 |
Hb_179118_010 |
0.0846365017 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 5 |
Hb_004590_010 |
0.1111342547 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 6 |
Hb_148914_020 |
0.121090827 |
- |
- |
PREDICTED: uncharacterized protein LOC105637821 [Jatropha curcas] |
| 7 |
Hb_005900_020 |
0.1252574342 |
- |
- |
- |
| 8 |
Hb_004631_030 |
0.1255685724 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 9 |
Hb_000002_230 |
0.1340302315 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |
| 10 |
Hb_002894_040 |
0.1387348889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_012395_150 |
0.1397693353 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator-like APRR7 [Jatropha curcas] |
| 12 |
Hb_011206_050 |
0.1459009046 |
- |
- |
hypothetical protein POPTR_0011s02840g [Populus trichocarpa] |
| 13 |
Hb_000203_150 |
0.1467666077 |
- |
- |
PREDICTED: uncharacterized protein LOC105116038 [Populus euphratica] |
| 14 |
Hb_001013_060 |
0.149948752 |
- |
- |
- |
| 15 |
Hb_000270_550 |
0.1502521898 |
- |
- |
- |
| 16 |
Hb_012879_010 |
0.1514226891 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001369_530 |
0.151970001 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 18 |
Hb_012516_030 |
0.1531535844 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 2 {ECO:0000303 |
| 19 |
Hb_004218_220 |
0.1541865993 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 20 |
Hb_001213_080 |
0.1550887303 |
- |
- |
PREDICTED: tubulin beta chain-like [Jatropha curcas] |