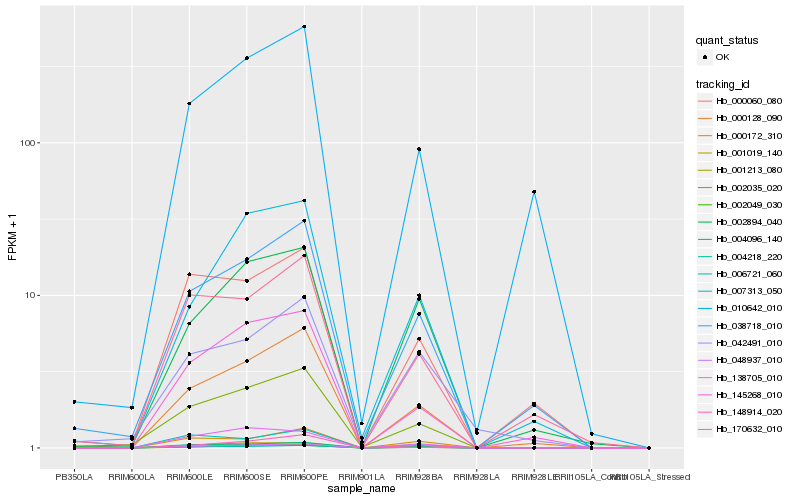
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002035_020 |
0.0 |
transcription factor |
TF Family: TRAF |
hypothetical protein B456_011G050200 [Gossypium raimondii] |
| 2 |
Hb_038718_010 |
0.1014987573 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 3 |
Hb_048937_010 |
0.1131483847 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1 [Nelumbo nucifera] |
| 4 |
Hb_000128_090 |
0.1165227703 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 5 |
Hb_004096_140 |
0.1167915128 |
- |
- |
Lipase, putative [Ricinus communis] |
| 6 |
Hb_001019_140 |
0.124130112 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_001213_080 |
0.1316509129 |
- |
- |
PREDICTED: tubulin beta chain-like [Jatropha curcas] |
| 8 |
Hb_148914_020 |
0.1360768414 |
- |
- |
PREDICTED: uncharacterized protein LOC105637821 [Jatropha curcas] |
| 9 |
Hb_145268_010 |
0.1368636527 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Jatropha curcas] |
| 10 |
Hb_170632_010 |
0.1455244001 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 11 |
Hb_004218_220 |
0.1460372802 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000172_310 |
0.1481584482 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 21 [Jatropha curcas] |
| 13 |
Hb_007313_050 |
0.1494747475 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 14 |
Hb_042491_010 |
0.1573936568 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 15 |
Hb_010642_010 |
0.1577027197 |
- |
- |
PREDICTED: aquaporin TIP2-1 [Jatropha curcas] |
| 16 |
Hb_002894_040 |
0.1614642958 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000060_080 |
0.1617207243 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 2 protein, putative [Ricinus communis] |
| 18 |
Hb_138705_010 |
0.1635193736 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 19 |
Hb_006721_060 |
0.1639272764 |
- |
- |
RnaseH (mitochondrion) [Malus domestica] |
| 20 |
Hb_002049_030 |
0.1664353199 |
- |
- |
PREDICTED: uncharacterized protein LOC105635940 isoform X3 [Jatropha curcas] |