| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
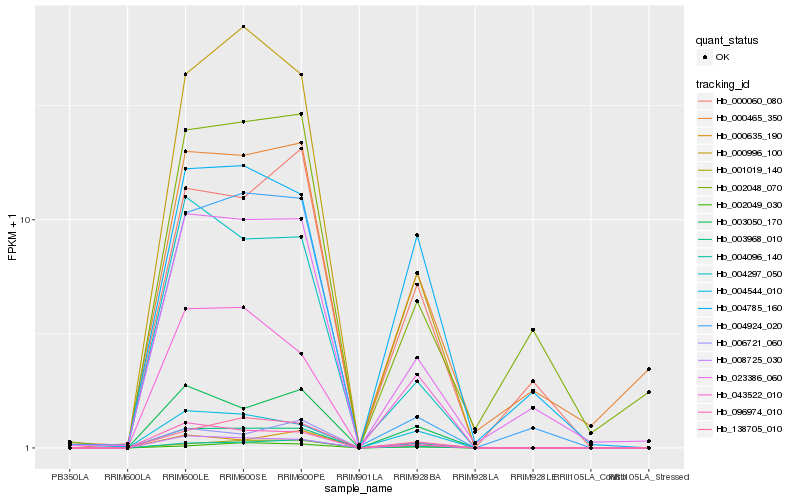
Hb_003968_010 |
0.0 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_008725_030 |
0.0701456049 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004096_140 |
0.0935872809 |
- |
- |
Lipase, putative [Ricinus communis] |
| 4 |
Hb_138705_010 |
0.1018980127 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 5 |
Hb_096974_010 |
0.1062542134 |
- |
- |
PREDICTED: uncharacterized protein LOC105123384 isoform X1 [Populus euphratica] |
| 6 |
Hb_003050_170 |
0.1138478648 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001019_140 |
0.1159747144 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_023386_060 |
0.1165397138 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 9 |
Hb_004297_050 |
0.1180768313 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_000996_100 |
0.1230780768 |
- |
- |
Nucleotide binding, putative [Theobroma cacao] |
| 11 |
Hb_006721_060 |
0.1313804513 |
- |
- |
RnaseH (mitochondrion) [Malus domestica] |
| 12 |
Hb_004544_010 |
0.1380940122 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Jatropha curcas] |
| 13 |
Hb_000060_080 |
0.1425659432 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 2 protein, putative [Ricinus communis] |
| 14 |
Hb_000465_350 |
0.1430435265 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 2 [Jatropha curcas] |
| 15 |
Hb_043522_010 |
0.1435384187 |
- |
- |
- |
| 16 |
Hb_000635_190 |
0.1449235105 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103322127 [Prunus mume] |
| 17 |
Hb_004785_160 |
0.1514545049 |
- |
- |
PREDICTED: U-box domain-containing protein 28-like [Jatropha curcas] |
| 18 |
Hb_004924_020 |
0.1523595934 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_002049_030 |
0.1539655916 |
- |
- |
PREDICTED: uncharacterized protein LOC105635940 isoform X3 [Jatropha curcas] |
| 20 |
Hb_002048_070 |
0.1550769514 |
- |
- |
hypothetical protein JCGZ_15146 [Jatropha curcas] |