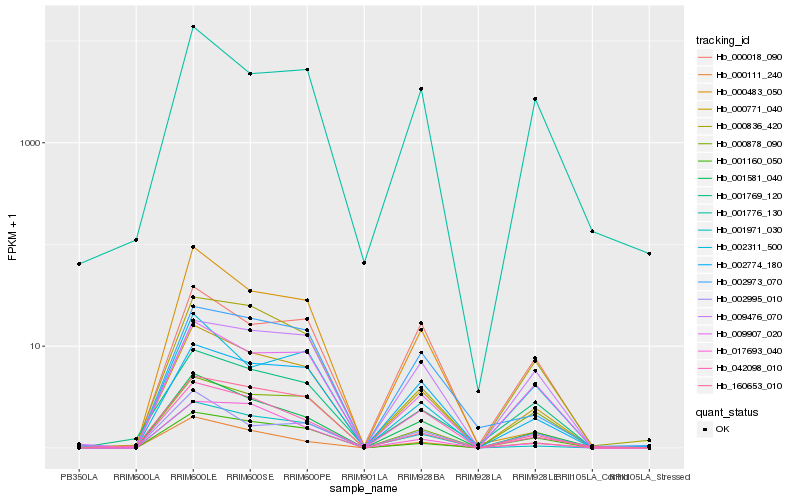
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000771_040 |
0.0 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like [Jatropha curcas] |
| 2 |
Hb_001581_040 |
0.0774976823 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_009907_020 |
0.0976408933 |
- |
- |
hypothetical protein CICLE_v10009323mg [Citrus clementina] |
| 4 |
Hb_001971_030 |
0.1042984897 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 5 |
Hb_002774_180 |
0.1093431379 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 6 |
Hb_000111_240 |
0.1150278935 |
- |
- |
PREDICTED: glutamate receptor 2.8-like [Elaeis guineensis] |
| 7 |
Hb_160653_010 |
0.1179720061 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 8 |
Hb_001160_050 |
0.1182757476 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO1 [Jatropha curcas] |
| 9 |
Hb_001769_120 |
0.1247417071 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 7A [Jatropha curcas] |
| 10 |
Hb_002995_010 |
0.129639122 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase [Jatropha curcas] |
| 11 |
Hb_000836_420 |
0.1356176539 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 12 |
Hb_042098_010 |
0.1387582876 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 13 |
Hb_001776_130 |
0.1404386038 |
- |
- |
metallothionein [Hevea brasiliensis] |
| 14 |
Hb_002973_070 |
0.1424515781 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 15 |
Hb_017693_040 |
0.144151972 |
- |
- |
tyrosine kinase family protein [Medicago truncatula] |
| 16 |
Hb_000483_050 |
0.14470291 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 17 |
Hb_000878_090 |
0.1464841179 |
- |
- |
sucrose-phosphatase family protein [Populus trichocarpa] |
| 18 |
Hb_000018_090 |
0.1470152518 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_002311_500 |
0.1481423935 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 20 |
Hb_009476_070 |
0.1482088404 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |