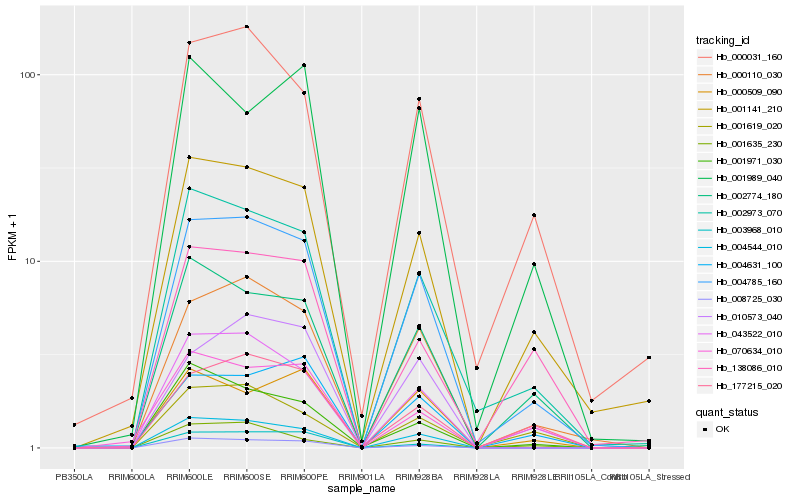
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004785_160 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 28-like [Jatropha curcas] |
| 2 |
Hb_000110_030 |
0.0752510171 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_002973_070 |
0.0876317666 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 4 |
Hb_004544_010 |
0.1000512379 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Jatropha curcas] |
| 5 |
Hb_001141_210 |
0.1002152111 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 6 |
Hb_000031_160 |
0.1118485628 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 7 |
Hb_043522_010 |
0.1139964819 |
- |
- |
- |
| 8 |
Hb_002774_180 |
0.1154005724 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 9 |
Hb_177215_020 |
0.126183462 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 10 |
Hb_008725_030 |
0.126380799 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_010573_040 |
0.1277005097 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal isoform X2 [Populus euphratica] |
| 12 |
Hb_001619_020 |
0.1403759749 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 13 |
Hb_004631_100 |
0.1426711489 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH96 [Jatropha curcas] |
| 14 |
Hb_000509_090 |
0.1453002848 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 15 |
Hb_070634_010 |
0.146106354 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 16 |
Hb_001971_030 |
0.1462181391 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 17 |
Hb_138086_010 |
0.1508532394 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 18 |
Hb_001989_040 |
0.1513483525 |
- |
- |
PREDICTED: neurofilament heavy polypeptide [Jatropha curcas] |
| 19 |
Hb_003968_010 |
0.1514545049 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_001635_230 |
0.1521494204 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |