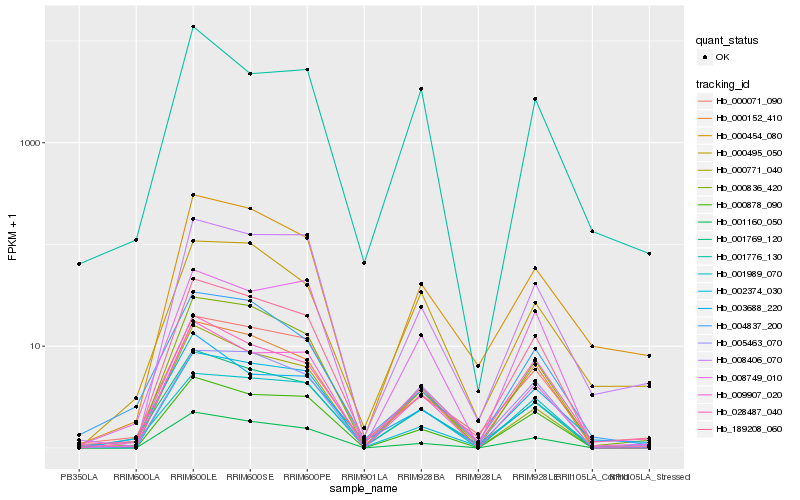
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001769_120 |
0.0 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 7A [Jatropha curcas] |
| 2 |
Hb_000152_410 |
0.0975803229 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 3 |
Hb_000071_090 |
0.0988495506 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |
| 4 |
Hb_009907_020 |
0.1071578755 |
- |
- |
hypothetical protein CICLE_v10009323mg [Citrus clementina] |
| 5 |
Hb_028487_040 |
0.1123023847 |
- |
- |
hypothetical protein POPTR_0010s11910g [Populus trichocarpa] |
| 6 |
Hb_001160_050 |
0.1179074311 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO1 [Jatropha curcas] |
| 7 |
Hb_000836_420 |
0.1180035923 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 8 |
Hb_000878_090 |
0.1211045064 |
- |
- |
sucrose-phosphatase family protein [Populus trichocarpa] |
| 9 |
Hb_189208_060 |
0.121926125 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 10 |
Hb_004837_200 |
0.1233438674 |
- |
- |
hypothetical protein POPTR_0001s26450g [Populus trichocarpa] |
| 11 |
Hb_000771_040 |
0.1247417071 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like [Jatropha curcas] |
| 12 |
Hb_000454_080 |
0.1280330953 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 13 |
Hb_003688_220 |
0.1280590756 |
- |
- |
unknown [Populus trichocarpa] |
| 14 |
Hb_008749_010 |
0.13436202 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 15 |
Hb_002374_030 |
0.1349997337 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001776_130 |
0.1355455733 |
- |
- |
metallothionein [Hevea brasiliensis] |
| 17 |
Hb_005463_070 |
0.1383841185 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 18 |
Hb_008406_070 |
0.1394851613 |
- |
- |
PREDICTED: probable protein phosphatase 2C 58 [Jatropha curcas] |
| 19 |
Hb_000495_050 |
0.1411427434 |
- |
- |
hypothetical protein JCGZ_25174 [Jatropha curcas] |
| 20 |
Hb_001989_070 |
0.1421629357 |
- |
- |
PREDICTED: uncharacterized calcium-binding protein At1g02270 [Jatropha curcas] |