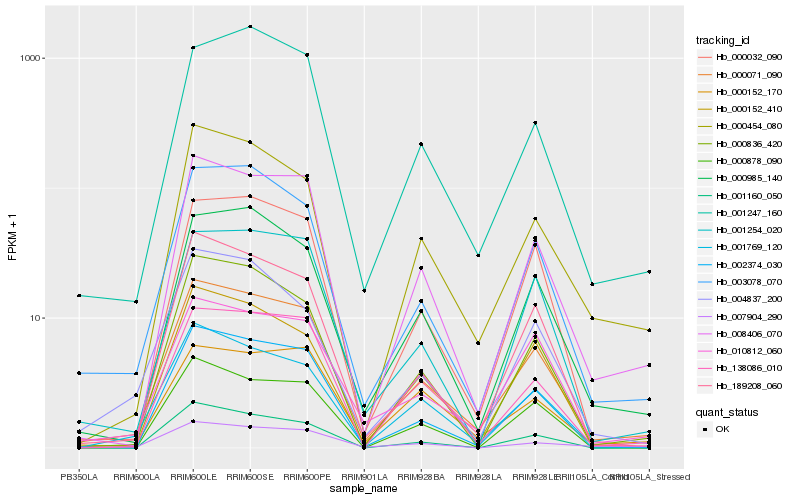
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000071_090 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |
| 2 |
Hb_008406_070 |
0.0830435072 |
- |
- |
PREDICTED: probable protein phosphatase 2C 58 [Jatropha curcas] |
| 3 |
Hb_003078_070 |
0.088732715 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At3g25290 [Jatropha curcas] |
| 4 |
Hb_002374_030 |
0.0907203651 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001769_120 |
0.0988495506 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 7A [Jatropha curcas] |
| 6 |
Hb_189208_060 |
0.1013043998 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 7 |
Hb_000985_140 |
0.1015310992 |
- |
- |
PREDICTED: uncharacterized protein LOC105643381 isoform X3 [Jatropha curcas] |
| 8 |
Hb_000454_080 |
0.1022769049 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 9 |
Hb_000836_420 |
0.105362289 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 10 |
Hb_010812_060 |
0.1065296705 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 11 |
Hb_000032_090 |
0.1083778416 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001254_020 |
0.1109959011 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 13 |
Hb_000152_410 |
0.1150350883 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 14 |
Hb_001247_160 |
0.1170338491 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 15 |
Hb_138086_010 |
0.1173115147 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 16 |
Hb_001160_050 |
0.120789349 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO1 [Jatropha curcas] |
| 17 |
Hb_007904_290 |
0.122780796 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 18 |
Hb_000878_090 |
0.1253040324 |
- |
- |
sucrose-phosphatase family protein [Populus trichocarpa] |
| 19 |
Hb_004837_200 |
0.1260808867 |
- |
- |
hypothetical protein POPTR_0001s26450g [Populus trichocarpa] |
| 20 |
Hb_000152_170 |
0.1283119181 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |