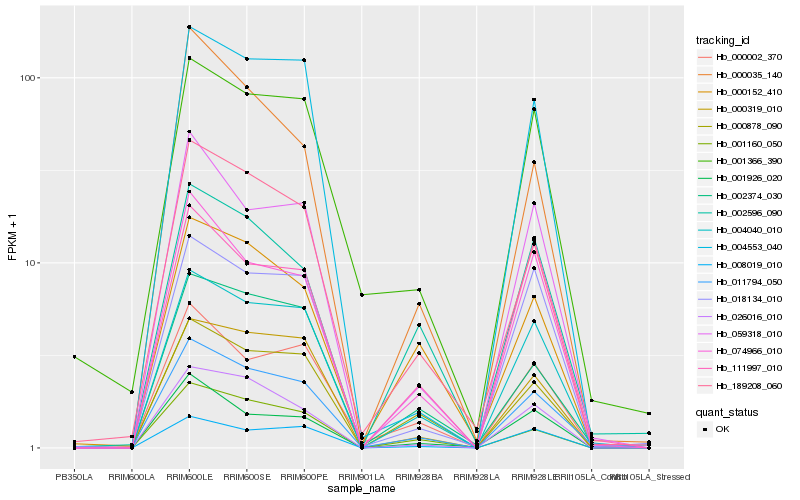
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011794_050 |
0.0 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_189208_060 |
0.0867652705 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 3 |
Hb_004040_010 |
0.0874898823 |
- |
- |
hypothetical protein POPTR_0019s14250g [Populus trichocarpa] |
| 4 |
Hb_002374_030 |
0.1077447138 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 5 |
Hb_059318_010 |
0.1079221566 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 6 |
Hb_026016_010 |
0.1119305758 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 7 |
Hb_074966_010 |
0.1120093087 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 8 |
Hb_000878_090 |
0.1127569452 |
- |
- |
sucrose-phosphatase family protein [Populus trichocarpa] |
| 9 |
Hb_000002_370 |
0.1140211564 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 10 |
Hb_001160_050 |
0.1146867644 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO1 [Jatropha curcas] |
| 11 |
Hb_000319_010 |
0.1154849922 |
- |
- |
hypothetical protein POPTR_0016s03600g [Populus trichocarpa] |
| 12 |
Hb_001366_390 |
0.1164194067 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 13 |
Hb_000152_410 |
0.1214702372 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 14 |
Hb_008019_010 |
0.1225854406 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 15 |
Hb_004553_040 |
0.1249469352 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 16 |
Hb_018134_010 |
0.1256751018 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 17 |
Hb_111997_010 |
0.127968373 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 18 |
Hb_002596_090 |
0.1298027407 |
- |
- |
PREDICTED: calcium-transporting ATPase 4, plasma membrane-type-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001926_020 |
0.132766162 |
- |
- |
PREDICTED: glutamate receptor 2.9-like [Jatropha curcas] |
| 20 |
Hb_000035_140 |
0.13283029 |
transcription factor |
TF Family: WRKY |
putative WRKY transcription factor family protein [Populus trichocarpa] |