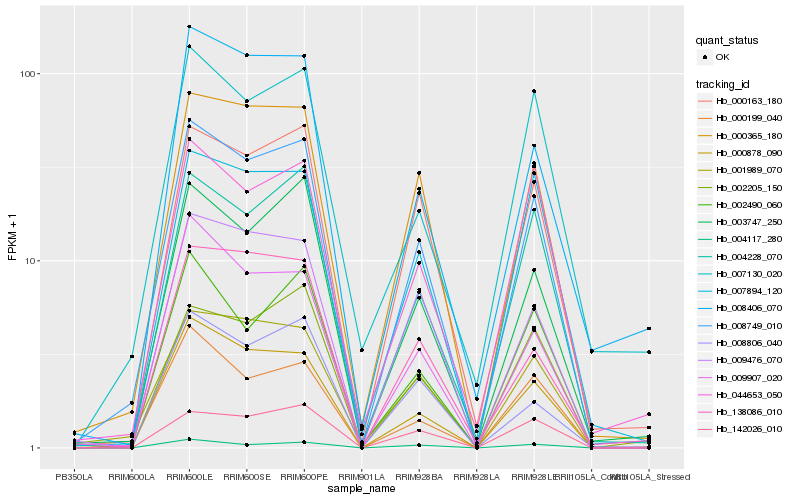
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008749_010 |
0.0 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 2 |
Hb_000365_180 |
0.0793886534 |
- |
- |
PREDICTED: protein LURP-one-related 8-like [Pyrus x bretschneideri] |
| 3 |
Hb_003747_250 |
0.087991307 |
- |
- |
PREDICTED: potassium transporter 7 isoform X2 [Elaeis guineensis] |
| 4 |
Hb_000878_090 |
0.0895128666 |
- |
- |
sucrose-phosphatase family protein [Populus trichocarpa] |
| 5 |
Hb_000163_180 |
0.095307444 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 6 |
Hb_004228_070 |
0.1007898175 |
- |
- |
PREDICTED: uncharacterized protein LOC103338699 [Prunus mume] |
| 7 |
Hb_001989_070 |
0.1028473801 |
- |
- |
PREDICTED: uncharacterized calcium-binding protein At1g02270 [Jatropha curcas] |
| 8 |
Hb_000199_040 |
0.1041299357 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_009476_070 |
0.110939485 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 10 |
Hb_009907_020 |
0.1130513337 |
- |
- |
hypothetical protein CICLE_v10009323mg [Citrus clementina] |
| 11 |
Hb_004117_280 |
0.1161009834 |
- |
- |
hypothetical protein JCGZ_00361 [Jatropha curcas] |
| 12 |
Hb_007894_120 |
0.1165410577 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_002490_060 |
0.1175192273 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 14 |
Hb_002205_150 |
0.1188656149 |
- |
- |
RING-H2 finger protein ATL3C, putative [Ricinus communis] |
| 15 |
Hb_008406_070 |
0.1195512942 |
- |
- |
PREDICTED: probable protein phosphatase 2C 58 [Jatropha curcas] |
| 16 |
Hb_142026_010 |
0.121594637 |
- |
- |
PREDICTED: probable mannitol dehydrogenase [Jatropha curcas] |
| 17 |
Hb_044653_050 |
0.121810339 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 18 |
Hb_008806_040 |
0.1233639276 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 19 |
Hb_138086_010 |
0.1233811556 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 20 |
Hb_007130_020 |
0.1255486298 |
- |
- |
- |